Locus Rank
7Gene
Id
127Gene Name
CHST12Duplicated
TrueMaternal
TrueGene Not Within Locus (Nearby
TrueGene Description
The protein encoded by this gene belongs to the sulfotransferase 2 family. It is localized to the golgi membrane, and catalyzes the transfer of sulfate to position 4 of the N-acetylgalactosamine (GalNAc) residue of chondroitin and desulfated dermatan sulfate. Chondroitin sulfate constitutes the predominant proteoglycan present in cartilage, and is distributed on the surfaces of many cells and extracellular matrices. Alternatively spliced transcript variants differing only in their 5' UTRs have been found for this gene. Mouse Phenotype
Additional Image
Allen Brain
Allen Regions
Zfin In Situ
Zfin Brain Areas
Published Zebrafish Pehnotype
Not In Allen Brain
FalseZFIN Link
NoneAllen Link
http://mouse.brain-map.org/gene/show/37594More Additional Images
Papers
http://www.ncbi.nlm.nih.gov/pubmed/25225104 on tenascin
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0121957
Locus Rank
Locus Rank
7Genes in Region
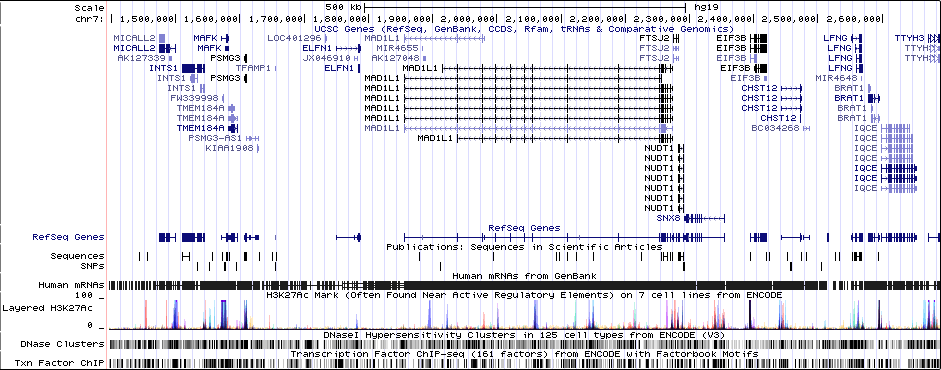
MAD1L1 (only gene in locus), and CHST12, SNX8, FTSJ2, EIF3B, NUDT1, and ELFN1 nearby locus.Associated Snps
GWAS Region (hg19)
chr7:1896096-2190096Genes Skipping
SNX8 - no brain expression, seems unlikely based on pubmed; NUDT1 - also not allen brain, and nothing about neurons in any paper; FTSJ2 - cell cycle, dna repair; EIF3B - essential roles in protein synthesisRicopili Plot Surrounding Area
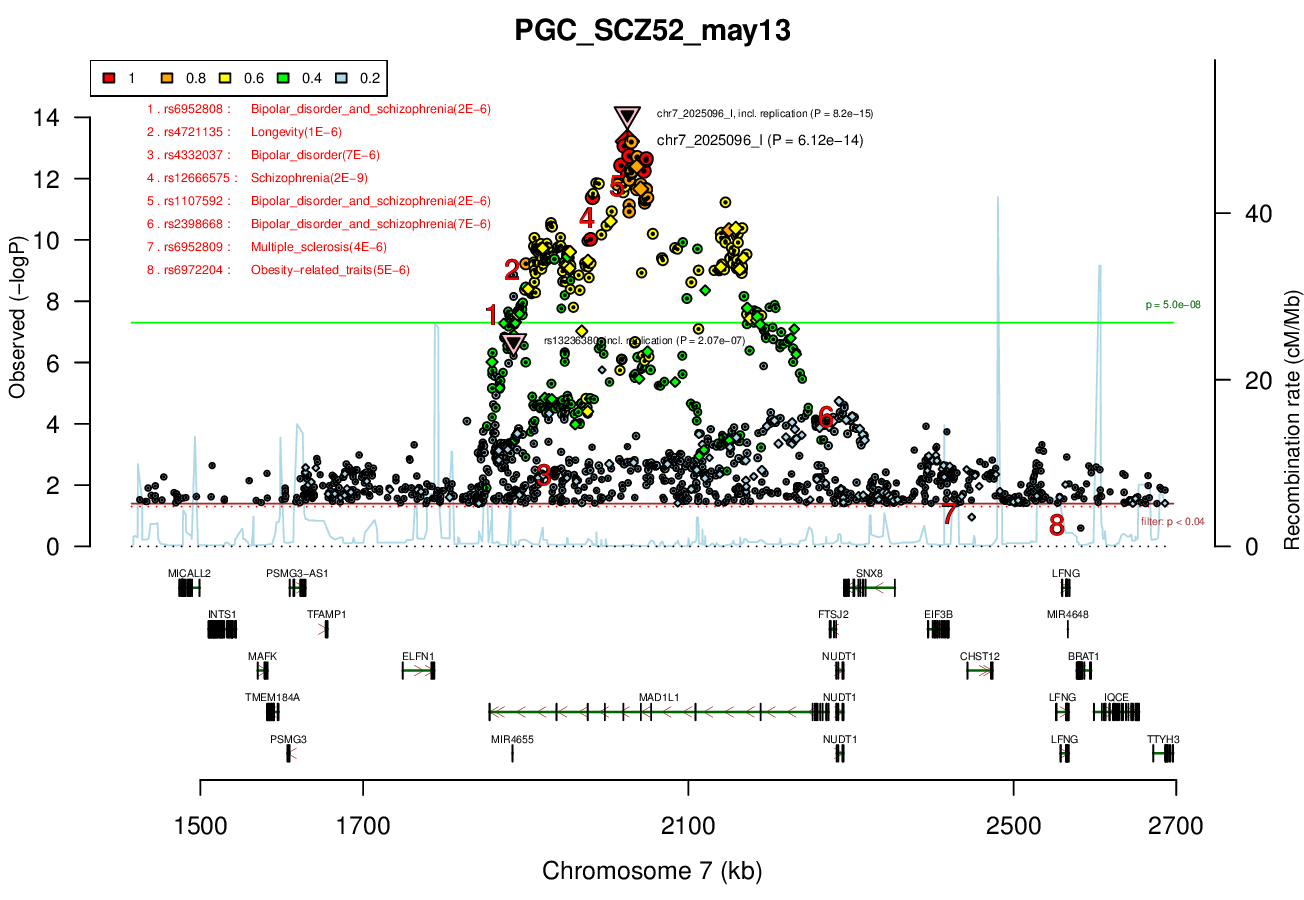
Distance On Each Side Of Locus In Above Plot
0.5 MBRicopili Plot
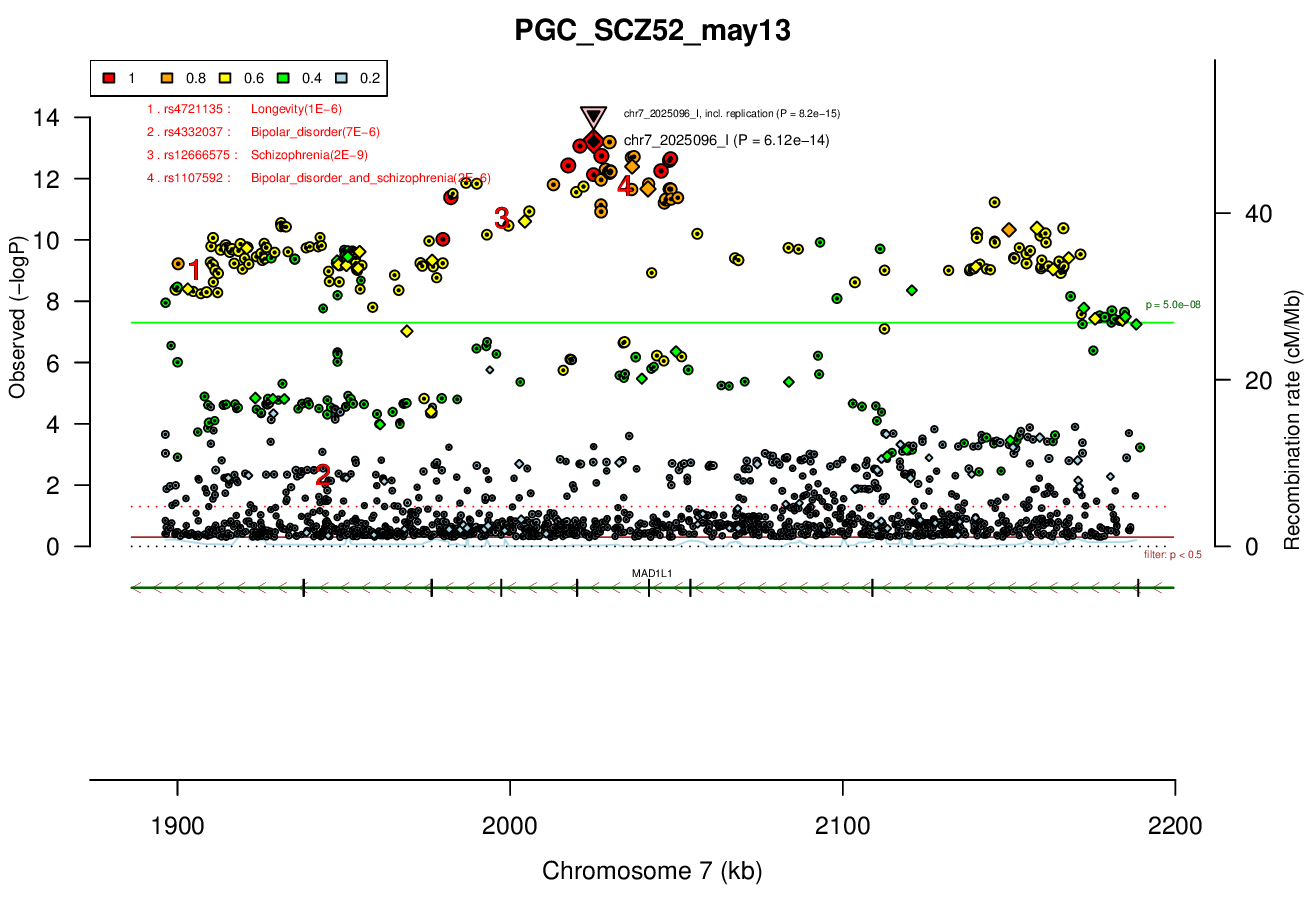
Gwas And Surrounding Region Pic
Gwas Region Pic
Sequence 1
Ensembl Gene Name
chst12aHarvard Allele
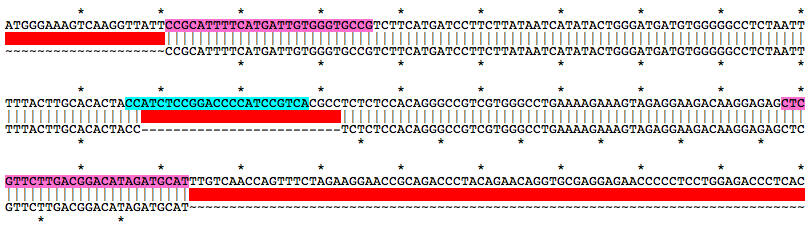
a252Mutation Area - WT DNA Sequence
CCGCATTTTCATGATTGTGGGTGCCGTCTTCATGATCCTTCTTATAATCATATACTGGGATGATGTGGGGGCCTCTAATTTTTACTTGCACACTACCATCTCCGGACCCCATCCGTCACGCCTCTCTCCACAGGGCCGTCGTGGGCCTGAAAAGAAAGTAGAGGAAGACAAGGAGAGCTCGTTCTTGACGGACATAGATGCATMutation Area - Mutant DNA Sequence
CCGCATTTTCATGATTGTGGGTGCCGTCTTCATGATCCTTCTTATAATCATATACTGGGATGATGTGGGGGCCTCTAATTTTTACTTGCACACTACCTCTCTCCACAGGGCCGTCGTGGGCCTGAAAAGAAAGTAGAGGAAGACAAGGAGAGCTCGTTCTTGACGGACATAGATGCATGuide RNA Target Sites
TGACGGATGGGGTCCGGAGATGGGenotyping Primers
f, CCGCATTTTCATGATTGTGGGTGCCG
r, ATGCATCTATGTCCGTCAAGAACGAG
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence
Allele
25bpDWT Genotyping Size
203WT Protein Sequence
MALSQSKSTKMGKSRLFRIFMIVGAVFMILLIIIYWDDVGASNFYLHTTISGPHPSRLSP
QGRRGPEKKVEEDKESSFLTDIDAFVNQFLEGTADPTEQVRGEPPPGDPHNQSSEKSDEK
FVPRREWKIHLSPIDPEKKQKQENRKQLIQDVCGNKSVFDFPGKNRTFDDIPNKELDHLI
VDDRHGIIYCYVPKVACTNWKRIMIVLSESLLVDGVPYQDPLDVPQELIHNSSLHFTFNK
FWKRYGKFSRHLMKIKLKKYTKFLFVRDPFVRLISAYRNKFEQENEDFYKRFALVMLKKY
SNYTDPPASVVDAFAAGIRPSFSNFVQYLLDPSTEKEMPFNEHWRQMYRLCHPCQINYDF
VGKLETLDEDAEHLLRILRVDNIVEFPESHRNRTVSSWEQDWFANIPLETRKELYRLYEA
DFKLFGYSKPEKLLHE-
Mutant Protein Sequence
MALSQSKSTKMGKSRLFRIFMIVGAVFMILLIIIYWDDVGASNFYLHTTSLHRAVVGLKRK-Mutant Genotyping Size
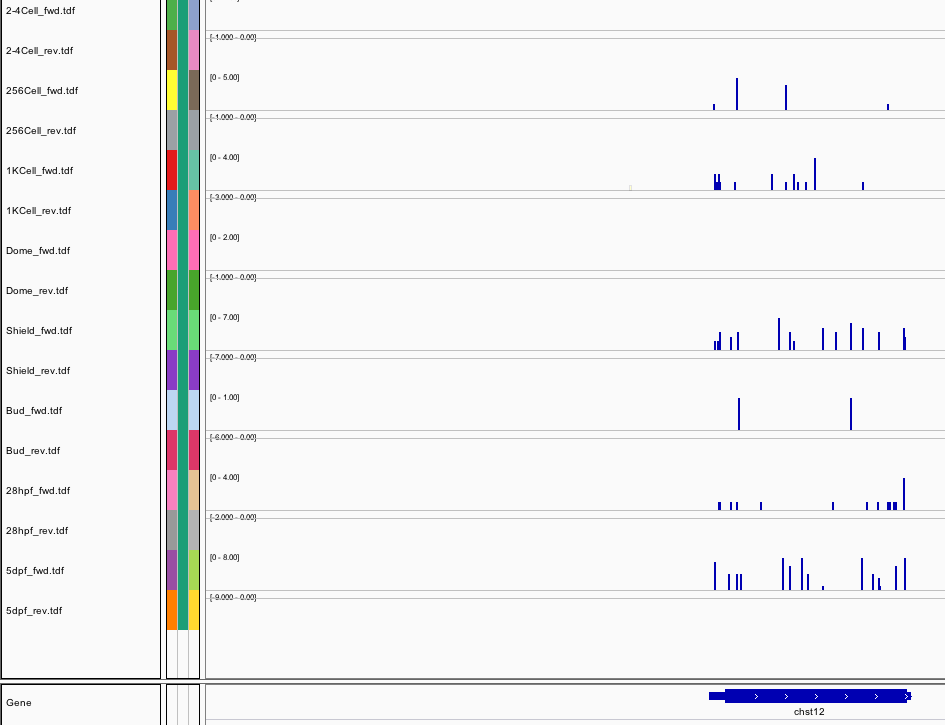
178Ribosome Profiling Development
Sequence 2
Ensembl Gene Name
chst12bHarvard Allele
a253Mutation Area - WT DNA Sequence
GATGGGACTTTTTATCTGCACGCAGCTTTATCAACACCTGCGCTCCCAGCAAGTGTTGTGTTCTCTGAACGTTGGACAAGCGAGGATTTGTTTATCTGGAAAGAGTCGAGCAGGTCAGTCAAACCAAACCTGACAATCATCTCAGCTCATCCAAACAATCTAACGCACATGTTCATGCAGTCTTCCTCCAAAAACCTCAAACTTCGTCAAGCACGTCGCAAACGTATAATAAAAGAACTTTGTCAAGCCAACAGCAGTTTGATTTTCCCTGGAAAATTTAGGACATTTGACCAAATTCCAAGCAAAGAGCTGGACCATCTCATTGTGGACGATCGTCACGGTGTGATTTACTGTTTTGTACCCAAGGTAGCTTGTACCAACTGGAAACGAATCATGATTGTGCTCAGCCAGAAGCTGAAGGCACCTGATGGAGCTCCATATCTGGATCCCCTTGACATACCTTCAGCTTTATCCCATAATGCTACTGTGCATTTGACATTTAACAAGTTTTGGAGGCTTTTTGGTCATTTTTCACGTCCTCTGATGCATCACAAGCTGAAGAACTACACTAAGTTCCTCTTTGTACGGGATCCTTTTGTACGACTGATTTCTGCTTTCCGGAACAAGTTTGCTCTGCCAAATGAGGATTTCTACAAACAGTTCGGCTCCACAATGCTTCAGCGTTATGCTAATATTTCTCAGCCCCCCACTTCGGCTCAAGAGGCCTTCAGTGCAGGCATCAGACTGTCCTTTACTCACTTCATTAAGTATCTGTTAGATCCACAGACTGAGAAGGAGAAGCCGTTMutation Area - Mutant DNA Sequence
GATGGGACTTTTTATCTGCACGCAGCTTTATCAACACCTGCGCTCCCAGCAAGTGTTGTGTTCTCTGACGTTGGACAAGCGAGGATTTGTTTATCTGGAAAGAGTCGAGCAGGTCAGTCAAACCAAACCGGACAATCATCTCAGCTCATCAAAACAATCTAACGCACATGTTCATGCAGTCTTCCTCCAAAAACCTCAAACTTCGTCAAGCGCGTCGCAAACGTATAATAAAAGAACTTTGTTTAGCCAACAGCAGTTTGAACTTCCCTGGAAAATTTAGGACATTTGACCAAATTCCAAGCAAAGAGCTGGACCAAGAGGCCTTCAGTGCAGGTATCAGACTGTCTTTTACTCACTTCATTAAGTATCTGTTAGATCCACAGACTGAGAAGGAGAAGCCGTTGuide RNA Target Sites
GTGTTGTGTTCTCTGAACGTTGG
GACGTGCTTGACGAAGTTTGAGG
AGAGCTGGACCATCTCATTGTGG
GAAGGCCTCTTGAGCCGAAGTGG
Genotyping Primers
f, GATGGGACTTTTTATCTGCACGCAGC
r, AACGGCTTCTCCTTCTCAGTCTGTGG
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence
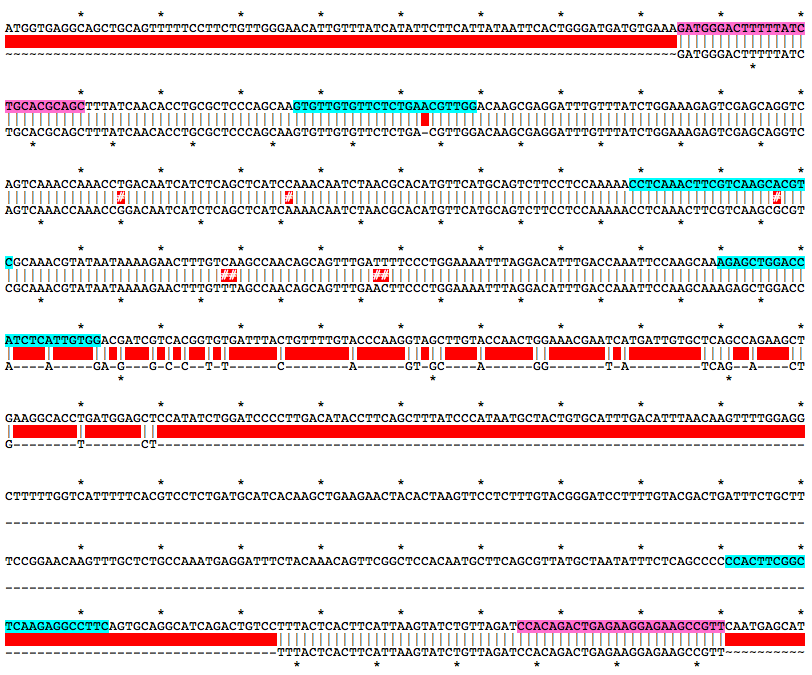
Allele
1bpD,402bpDWT Genotyping Size
806WT Protein Sequence
MVRQLQFFLLLGTLFIIFFIIIHWDDVKDGTFYLHAALSTPALPASVVFSERWTSEDLFI
WKESSRSVKPNLTIISAHPNNLTHMFMQSSSKNLKLRQARRKRIIKELCQANSSLIFPGK
FRTFDQIPSKELDHLIVDDRHGVIYCFVPKVACTNWKRIMIVLSQKLKAPDGAPYLDPLD
IPSALSHNATVHLTFNKFWRLFGHFSRPLMHHKLKNYTKFLFVRDPFVRLISAFRNKFAL
PNEDFYKQFGSTMLQRYANISQPPTSAQEAFSAGIRLSFTHFIKYLLDPQTEKEKPFNEH
WQQMHRLCHPCQIDYDFVGKLETLHEDTEHLLKILGLSNQIRFPPGYRNRTAVKWEREWF
ANVSVADRRKLYSLYEADFRLFGYNKPETLF-
Mutant Protein Sequence
MVRQLQFFLLLGTLFIIFFIIIHWDDVKDGTFYLHAALSTPALPASVVFSERWTSEDLFIWKESSRS
VKPNRTIISAHQNNLTHMFMQSSSKNLKLRQARRKRIIKELCLANSSLNFPGKFRTFDQIPSKELDQE
AFSAGIRLSFTHFIKYLLDPQTEKEKPFNEHWQQMHRLCHPCQIDYDFVGKLETLHEDTEHLLKILGL
SNQIRFPPGYRNRTAVKWEREWFANVSVADRRKLYSLYEADFRLFGYNKPETLF-
Mutant Genotyping Size
403Behavior Data
Behavior Data Summary
NoneBehavior Data Description 1
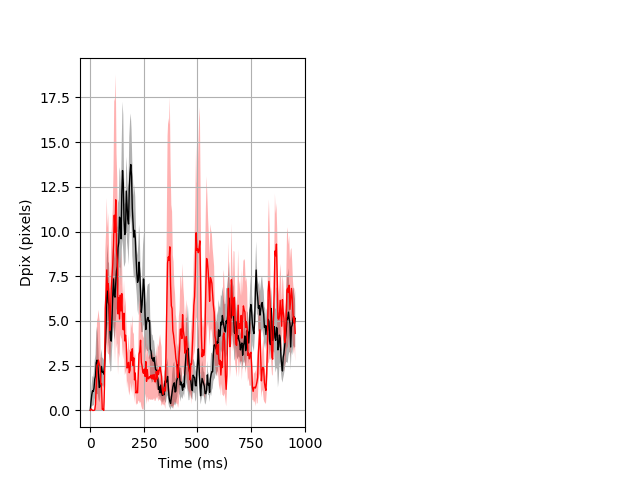
Dark flash block 1 start Merged section Pvalue = not significant 14 hethet vs 8 homhom ribgraph_mean_ribbon_fullboutdata_dpix_a0darkflash103.pngBehavior Data Graph 1
Behavior Data Description 2
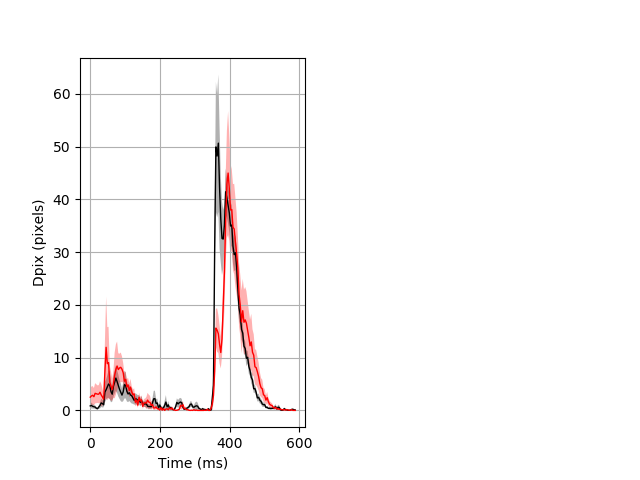
Day all prepulse tap Merged section Pvalue = 0.0132 14 hethet vs 8 homhom ribgraph_mean_ribbon_fullboutdata_dpix_dayprepulseinhibition100d.pngBehavior Data Graph 2
Behavior Data Description 3
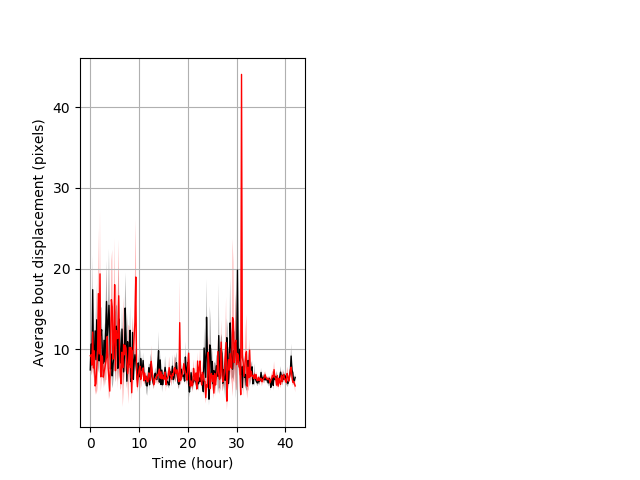
Features of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 14 hethet vs 8 homhom ribgraph_mean_ribbonbout_aveboutdisp_10min_combo.pngBehavior Data Graph 3
Behavior Data Description 4
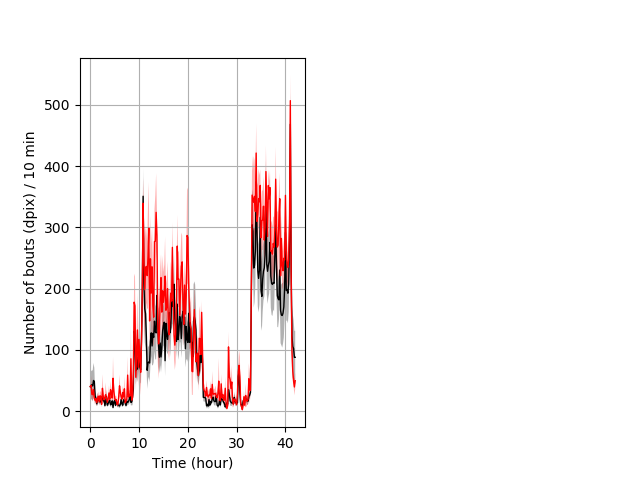
Frequency of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 14 hethet vs 8 homhom ribgraph_mean_ribbonbout_dpixnumberofbouts_10min_combo.pngBehavior Data Graph 4
Behavior Data Description 5
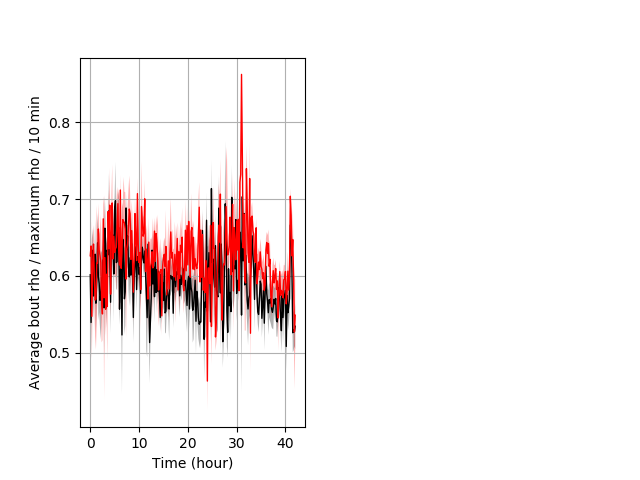
Location in well entire protocol Graph Pvalue = 0.0478 Merged section Pvalue = not significant 14 hethet vs 8 homhom ribgraph_mean_ribbonbout_averhofrac_10min_combo.pngBehavior Data Graph 5
Behavior Data Description 6
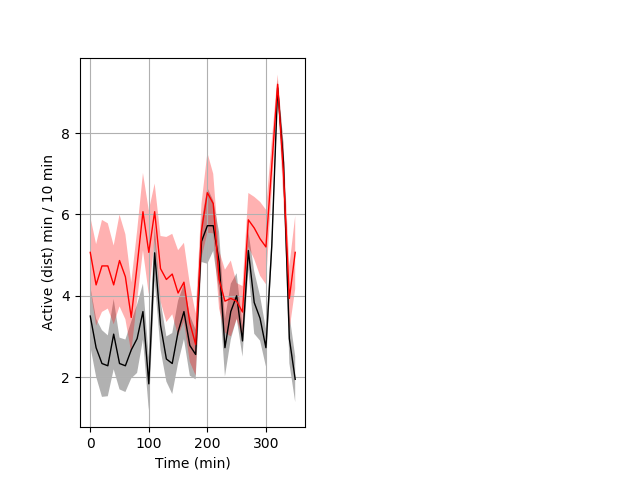
Frequency of movement Graph Pvalue = 0.014 18 hethet vs 15 hethom ribgraph_mean_ribbon_distminper_10min_day2nightstim.pngBehavior Data Graph 6
Behavior Data Description 7
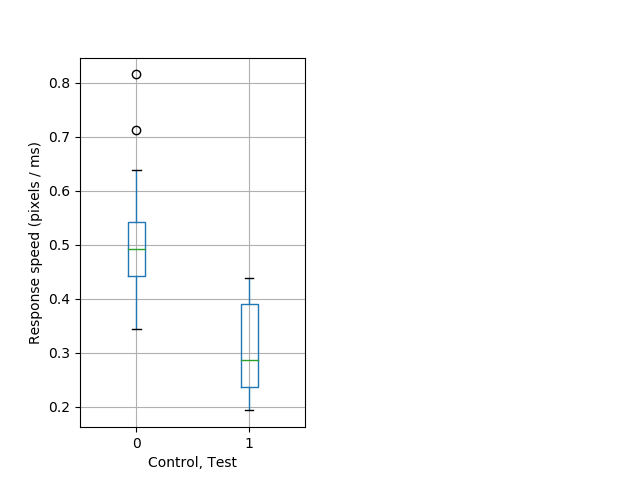
Strong tap Graph Pvalue = 0.00134 14 hethet vs 8 homhom boxgraph_pd__ribgraph_mean_ribbon_speedresponse_dist_nightprepulseinhibition102.pngBehavior Data Graph 7
Behavior Data Description 8
Tap habituation Graph Pvalue = 0.00459 18 hethet vs 9 homhom boxgraph_pd__ribgraph_mean_ribbon_displacement_dist_cdaytaphab1.pngBehavior Data Graph 8
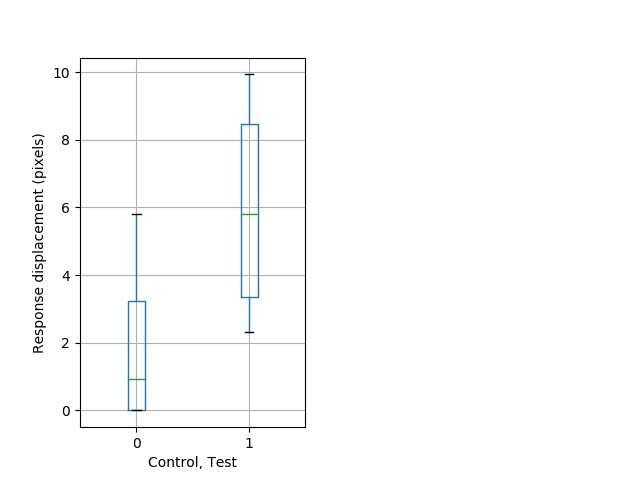
Behavior Data Description 9
NoneBehavior Data Graph 9
NoneBehavior Data Description 10
NoneBehavior Data Graph 10
NoneBehavior Data Description 11
NoneBehavior Data Graph 11
NoneBehavior Data Description 12
NoneBehavior Data Graph 12
NoneBehavior Data Description 13
NoneBehavior Data Graph 13
NoneBehavior Data Description 14
NoneBehavior Data Graph 14
None