Locus Rank
7Gene
Id
129Gene Name
ELFN1Duplicated
TrueMaternal
FalseGene Not Within Locus (Nearby
TrueGene Description
Postsynaptic protein that regulates circuit dynamics in the central nervous system by modulating the temporal dynamics of interneuron recruitment. Mouse Phenotype
loss of Elfn1 results in hyperactivity and sensory-triggered epileptic seizures in miceAdditional Image
Allen Brain
Allen Regions
Zfin In Situ
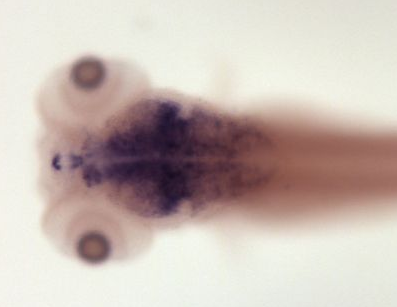
Zfin Brain Areas
elf1b: diencephalon, habenula, hindbrain, midbrain, midbrain hindbrain boundary, neuron, optic tectum, tegmentum, telencephalonPublished Zebrafish Pehnotype
Not In Allen Brain
FalseZFIN Link
http://zfin.org/search?q=elfn1&fq=category%3A%22Gene+%2F+Transcript%22&category=Gene+%2F+Transcript&fq=type%3A%22Gene%22Allen Link
http://mouse.brain-map.org/gene/show/89064More Additional Images
Papers
http://www.ncbi.nlm.nih.gov/pubmed/25047565
http://www.ncbi.nlm.nih.gov/pubmed/24312227
http://www.ncbi.nlm.nih.gov/pubmed/23042292
Locus Rank
Locus Rank
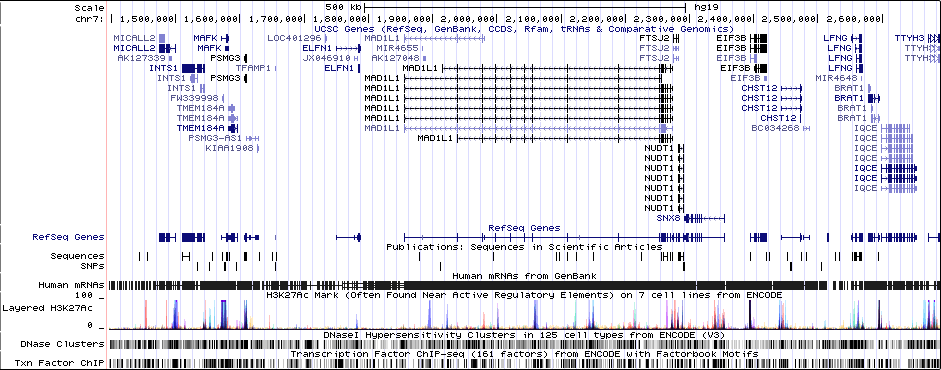
7Genes in Region
MAD1L1 (only gene in locus), and CHST12, SNX8, FTSJ2, EIF3B, NUDT1, and ELFN1 nearby locus.Associated Snps
GWAS Region (hg19)
chr7:1896096-2190096Genes Skipping
SNX8 - no brain expression, seems unlikely based on pubmed; NUDT1 - also not allen brain, and nothing about neurons in any paper; FTSJ2 - cell cycle, dna repair; EIF3B - essential roles in protein synthesisRicopili Plot Surrounding Area
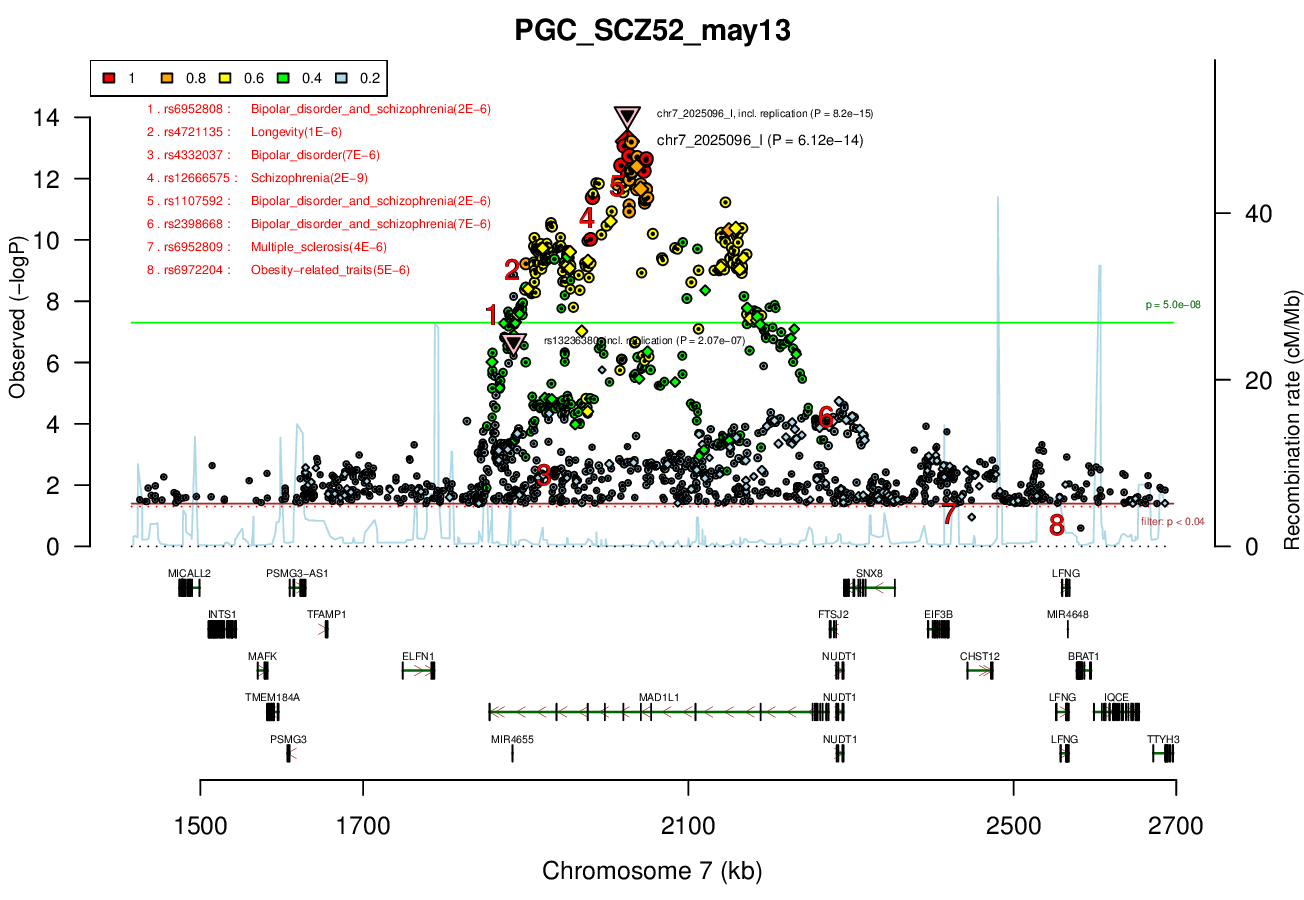
Distance On Each Side Of Locus In Above Plot
0.5 MBRicopili Plot
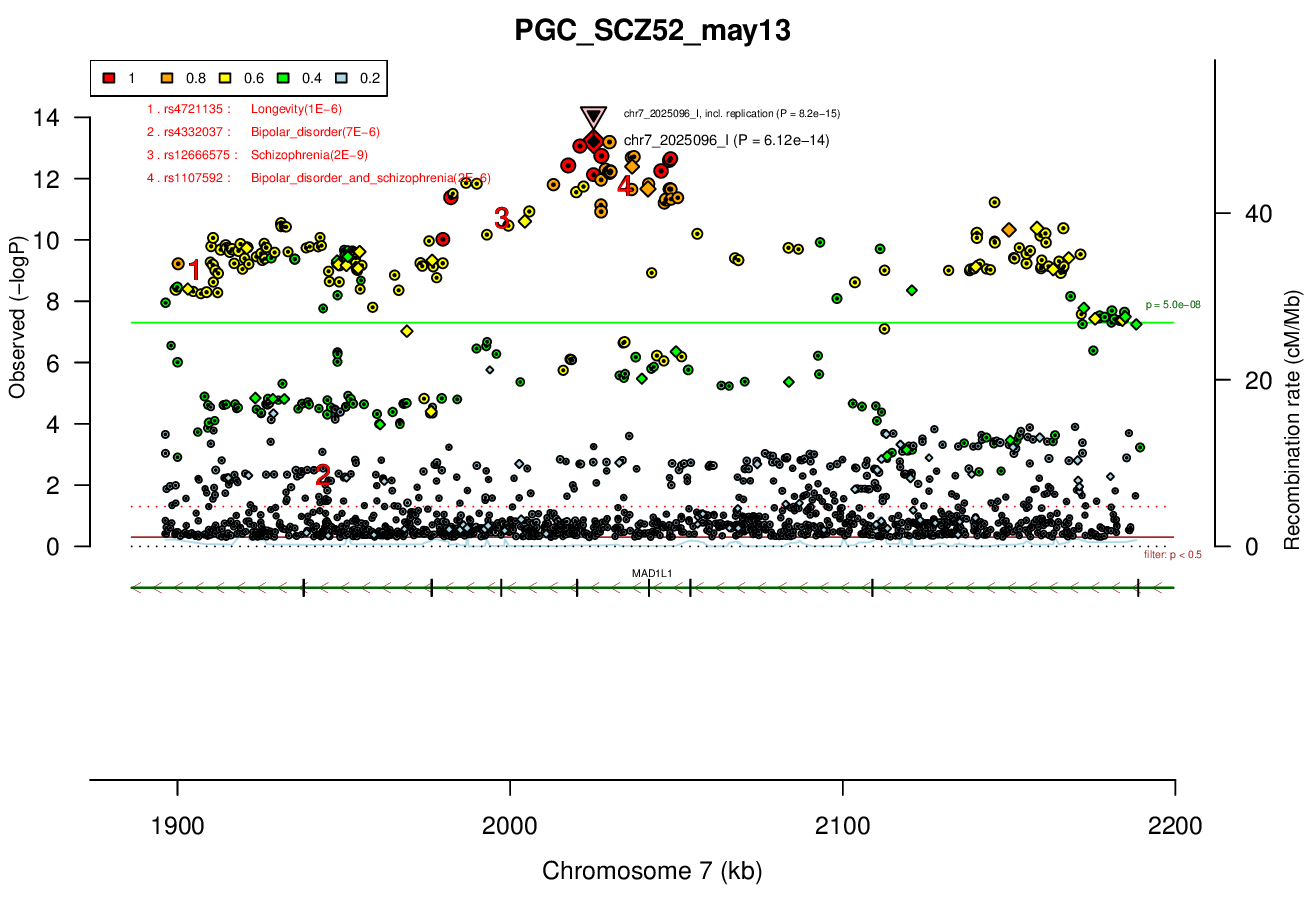
Gwas And Surrounding Region Pic
Gwas Region Pic
Sequence 1
Ensembl Gene Name
elfn1a, ELFN1 (1 of many)Harvard Allele
a216Mutation Area - WT DNA Sequence
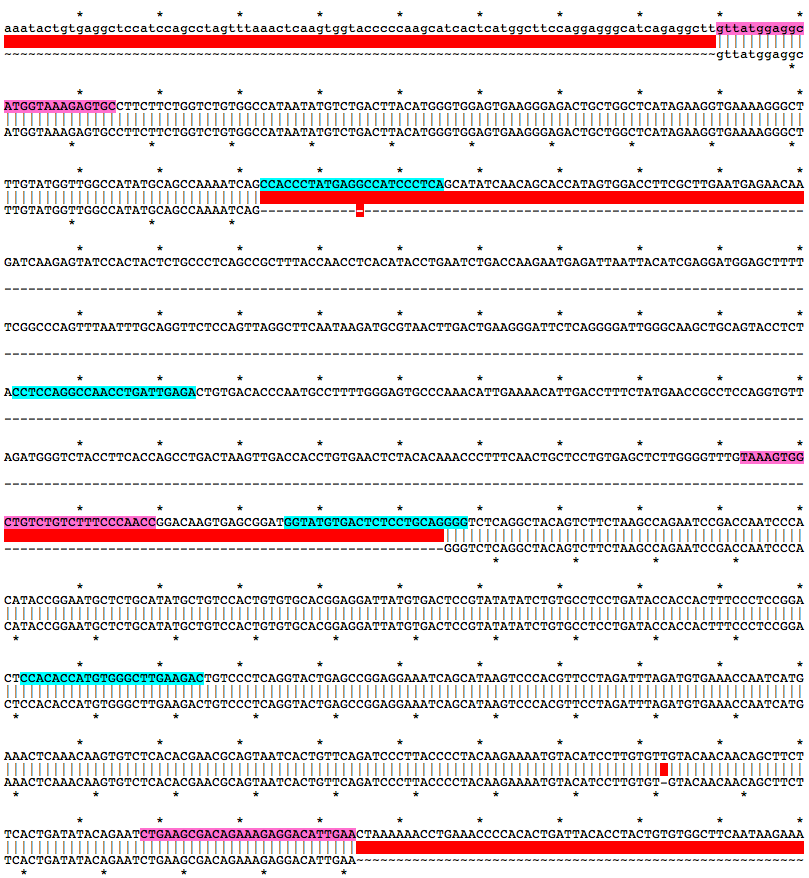
gttatggaggcATGGTAAAGAGTGCCTTCTTCTGGTCTGTGGCCATAATATGTCTGACTTACATGGGTGGAGTGAAGGGAGACTGCTGGCTCATAGAAGGTGAAAAGGGCTTTGTATGGTTGGCCATATGCAGCCAAAATCAGCCACCCTATGAGGCCATCCCTCAGCATATCAACAGCACCATAGTGGACCTTCGCTTGAATGAGAACAAGATCAAGAGTATCCACTACTCTGCCCTCAGCCGCTTTACCAACCTCACATACCTGAATCTGACCAAGAATGAGATTAATTACATCGAGGATGGAGCTTTTTCGGCCCAGTTTAATTTGCAGGTTCTCCAGTTAGGCTTCAATAAGATGCGTAACTTGACTGAAGGGATTCTCAGGGGATTGGGCAAGCTGCAGTACCTCTACCTCCAGGCCAACCTGATTGAGACTGTGACACCCAATGCCTTTTGGGAGTGCCCAAACATTGAAAACATTGACCTTTCTATGAACCGCCTCCAGGTGTTAGATGGGTCTACCTTCACCAGCCTGACTAAGTTGACCACCTGTGAACTCTACACAAACCCTTTCAACTGCTCCTGTGAGCTCTTGGGGTTTGTAAAGTGGCTGTCTGTCTTTCCCAACCGGACAAGTGAGCGGATGGTATGTGACTCTCCTGCAGGGGTCTCAGGCTACAGTCTTCTAAGCCAGAATCCGACCAATCCCACATACCGGAATGCTCTGCATATGCTGTCCACTGTGTGCACGGAGGATTATGTGACTCCGTATATATCTGTGCCTCCTGATACCACCACTTTCCCTCCGGACTCCACACCATGTGGGCTTGAAGACTGTCCCTCAGGTACTGAGCCGGAGGAAATCAGCATAAGTCCCACGTTCCTAGATTTAGATGTGAAACCAATCATGAAACTCAAACAAGTGTCTCACACGAACGCAGTAATCACTGTTCAGATCCCTTACCCCTACAAGAAAATGTACATCCTTGTGTTGTACAACAACAGCTTCTTCACTGATATACAGAATCTGAAGCGACAGAAAGAGGACATTGAAMutation Area - Mutant DNA Sequence
gttatggaggcATGGTAAAGAGTGCCTTCTTCTGGTCTGTGGCCATAATATGTCTGACTTACATGGGTGGAGTGAAGGGAGACTGCTGGCTCATAGAAGGTGAAAAGGGCTTTGTATGGTTGGCCATATGCAGCCAAAATCAGGGGTCTCAGGCTACAGTCTTCTAAGCCAGAATCCGACCAATCCCACATACCGGAATGCTCTGCATATGCTGTCCACTGTGTGCACGGAGGATTATGTGACTCCGTATATATCTGTGCCTCCTGATACCACCACTTTCCCTCCGGACTCCACACCATGTGGGCTTGAAGACTGTCCCTCAGGTACTGAGCCGGAGGAAATCAGCATAAGTCCCACGTTCCTAGATTTAGATGTGAAACCAATCATGAAACTCAAACAAGTGTCTCACACGAACGCAGTAATCACTGTTCAGATCCCTTACCCCTACAAGAAAATGTACATCCTTGTGTGTACAACAACAGCTTCTTCACTGATATACAGAATCTGAAGCGACAGAAAGAGGACATTGAAGuide RNA Target Sites
TGAGGGATGGCCTCATAGGGTGG
TCTCAATCAGGTTGGCCTGGAGG
GGTATGTGACTCTCCTGCAGGGG
GTCTTCAAGCCCACATGGTGTGG
Genotyping Primers
f, gttatggaggcATGGTAAAGAGTGC
r, TTCAATGTCCTCTTTCTGTCGCTTCAG
wt_f, TAAAGTGGCTGTCTGTCTTTCCCAACC
PCR with mix of all three primers.
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence
Allele
524bpDWT Genotyping Size
452WT Protein Sequence
MASRRASEACYGGMVKSAFFWSVAIICLTYMGGVKGDCWLIEGEKGFVWLAICSQNQPPY
EAIPQHINSTIVDLRLNENKIKSIHYSALSRFTNLTYLNLTKNEINYIEDGAFSAQFNLQ
VLQLGFNKMRNLTEGILRGLGKLQYLYLQANLIETVTPNAFWECPNIENIDLSMNRLQVL
DGSTFTSLTKLTTCELYTNPFNCSCELLGFVKWLSVFPNRTSERMVCDSPAGVSGYSLLS
QNPTNPTYRNALHMLSTVCTEDYVTPYISVPPDTTTFPPDSTPCGLEDCPSGTEPEEISI
SPTFLDLDVKPIMKLKQVSHTNAVITVQIPYPYKKMYILVLYNNSFFTDIQNLKRQKEDI
ELKNLKPHTDYTYCVASIRNSLRFNHTCLTLSTGPKNGKERVASNATATHYIMTILGCLF
SMVIVLGIVYYCLRKKRQQDEKHKKAGSLKKNIIELKYGGELEGGTISRMSQKQMMAGES
MTRMPYLPSGSEMEQYKLQDISDTPKMAKGNYMEVRTGEHPDRRDCEMSMAGNSQGSVAE
ISTIAKEVDKVNQIINNCIDALKSESTSFQGVKSGAVSTAEPQLVLISEQPQSKSGFLSP
VYKDSYHHSLQRHHASDASPKRPSTATGGSIRSPRPYRSEGSYKSESKYIEKTSPTGETI
LTITPAAAILRAEAEKIRQYSEHRHSYPDAHQIEELEGPESRKTSILEPLTRPRARDLAY
SQLSPQYHNLSYSSSPEYYCKPSHSIWERFKLHRKRHKDEEYMAAGHALRKKVQFAKDED
LHDILDYWKGVSAQQKS-
Mutant Protein Sequence
MASRRASEACYGGMVKSAFFWSVAIICLTYMGGVKGDCWLIEGEKGFVWLAICSQNQGSQATVF-In Situ Probe Sequence
GCCTTCTTCTGGTCTGTGGCCATAATATGTCTGACTTACATGGGTGGAGTGAAGGGAGACTGCTGGCTCATAGAAGGTGAAAAGGGCTTTGTATGGTTGGCCATATGCAGCCAAAATCAGCCACCCTATGAGGCCATCCCTCAGCATATCAACAGCACCATAGTGGACCTTCGCTTGAATGAGAACAAGATCAAGAGTATCCACTACTCTGCCCTCAGCCGCTTTACCAACCTCACATACCTGAATCTGACCAAGAATGAGATTAATTACATCGAGGATGGAGCTTTTTCGGCCCAGTTTAATTTGCAGGTTCTCCAGTTAGGCTTCAATAAGATGCGTAACTTGACTGAAGGGATTCTCAGGGGATTGGGCAAGCTGCAGTACCTCTACCTCCAGGCCAACCTGATTGAGACTGTGACACCCAATGCCTTTTGGGAGTGCCCAAACATTGAAAACATTGACCTTTCTATGAACCGCCTCCAGGTGTTAGATGGGTCTACCTTCACCAGCCTGACTAAGTTGACCACCTGTGAACTCTACACAAACCCTTTCAACTGCTCCTGTGAGCTCTTGGGGTTTGTAAAGTGGCTGTCTGTCTTTCCCAACCGGACAAGTGAGCGGATGGTATGTGACTCTCCTGCAGGGGTCTCAGGCTACAGTCTTCTAAGCCAGAATCCGACCAATCCCACATACCGGAATGCTCTGCATATGCTGTCCACTGTGTGCACGGAGGATTATGTGACTCCGTATATATCTGTGCCTCCTGATACCACCACTTTCCCTCCGGACTCCACACCATGTGGGCTTGAAGACTGTCCCTCAGGTACTGAGCCGGAGGAAATCAGCATAAGTCCCACGTTCCTAGATTTAGATGTGAAACCAATCATGAAACTCAAACAAGTGTCTCACACGAACGCAGTAATCACTGTTCAGATCCCTTACCCCTACAAGAAAATGTACATCCTTGTGTTGTACAACAACAGCTTCTTCACTGATATACAGAATCTGAAGCGACAGAAAGAGGMutant Genotyping Size
531Ribosome Profiling Development
Sequence 2
Ensembl Gene Name
elfn1bHarvard Allele
a217Mutation Area - WT DNA Sequence
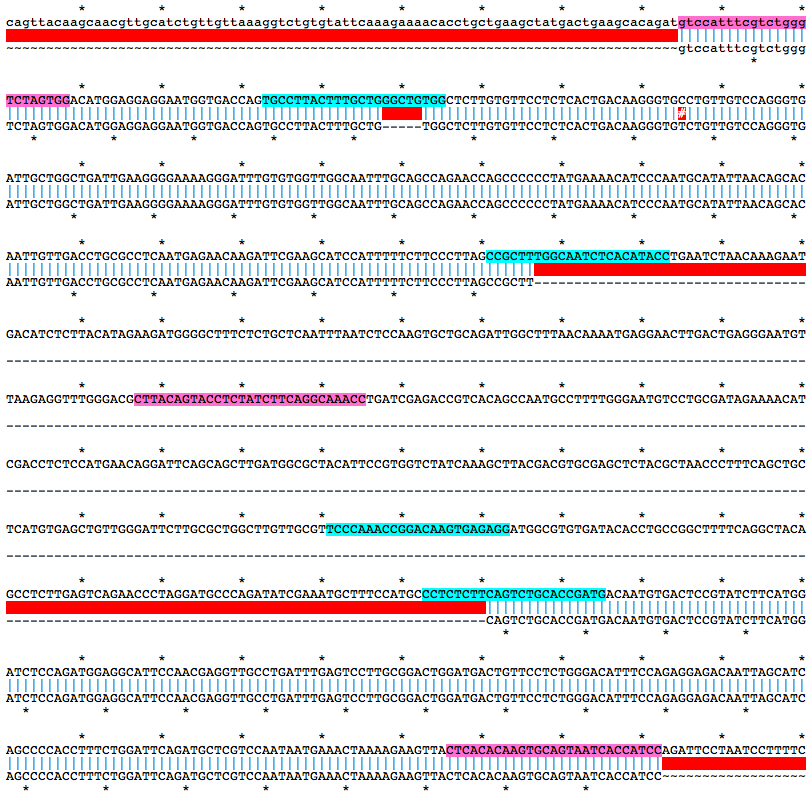
gtccatttcgtctgggTCTAGTGGACATGGAGGAGGAATGGTGACCAGTGCCTTACTTTGCTGGGCTGTGGCTCTTGTGTTCCTCTCACTGACAAGGGTGCCTGTTGTCCAGGGTGATTGCTGGCTGATTGAAGGGGAAAAGGGATTTGTGTGGTTGGCAATTTGCAGCCAGAACCAGCCCCCCTATGAAAACATCCCAATGCATATTAACAGCACAATTGTTGACCTGCGCCTCAATGAGAACAAGATTCGAAGCATCCATTTTTCTTCCCTTAGCCGCTTTGGCAATCTCACATACCTGAATCTAACAAAGAATGACATCTCTTACATAGAAGATGGGGCTTTCTCTGCTCAATTTAATCTCCAAGTGCTGCAGATTGGCTTTAACAAAATGAGGAACTTGACTGAGGGAATGTTAAGAGGTTTGGGACGCTTACAGTACCTCTATCTTCAGGCAAACCTGATCGAGACCGTCACAGCCAATGCCTTTTGGGAATGTCCTGCGATAGAAAACATCGACCTCTCCATGAACAGGATTCAGCAGCTTGATGGCGCTACATTCCGTGGTCTATCAAAGCTTACGACGTGCGAGCTCTACGCTAACCCTTTCAGCTGCTCATGTGAGCTGTTGGGATTCTTGCGCTGGCTTGTTGCGTTCCCAAACCGGACAAGTGAGAGGATGGCGTGTGATACACCTGCCGGCTTTTCAGGCTACAGCCTCTTGAGTCAGAACCCTAGGATGCCCAGATATCGAAATGCTTTCCATGCCCTCTCTTCAGTCTGCACCGATGACAATGTGACTCCGTATCTTCATGGATCTCCAGATGGAGGCATTCCAACGAGGTTGCCTGATTTGAGTCCTTGCGGACTGGATGACTGTTCCTCTGGGACATTTCCAGAGGAGACAATTAGCATCAGCCCCACCTTTCTGGATTCAGATGCTCGTCCAATAATGAAACTAAAAGAAGTTACTCACACAAGTGCAGTAATCACCATCCMutation Area - Mutant DNA Sequence
gtccatttcgtctgggTCTAGTGGACATGGAGGAGGAATGGTGACCAGTGCCTTACTTTGCTGTGGCTCTTGTGTTCCTCTCACTGACAAGGGTGTCTGTTGTCCAGGGTGATTGCTGGCTGATTGAAGGGGAAAAGGGATTTGTGTGGTTGGCAATTTGCAGCCAGAACCAGCCCCCCTATGAAAACATCCCAATGCATATTAACAGCACAATTGTTGACCTGCGCCTCAATGAGAACAAGATTCGAAGCATCCATTTTTCTTCCCTTAGCCGCTTCAGTCTGCACCGATGACAATGTGACTCCGTATCTTCATGGATCTCCAGATGGAGGCATTCCAACGAGGTTGCCTGATTTGAGTCCTTGCGGACTGGATGACTGTTCCTCTGGGACATTTCCAGAGGAGACAATTAGCATCAGCCCCACCTTTCTGGATTCAGATGCTCGTCCAATAATGAAACTAAAAGAAGTTACTCACACAAGTGCAGTAATCACCATCCGuide RNA Target Sites
TGCCTTACTTTGCTGGGCTGTGG
GGTATGTGAGATTGCCAAAGCGG
TCCCAAACCGGACAAGTGAGAGG
CATCGGTGCAGACTGAAGAGAGG
Genotyping Primers
f, gtccatttcgtctgggTCTAGTGG
r, GGATGGTGATTACTGCACTTGTGTGAG
wt_f, CTTACAGTACCTCTATCTTCAGGCAAACC
Mix of three primers works for genotyping.
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence
Allele
6bpd,494bpdWT Genotyping Size
566 (with wt_f)WT Protein Sequence
MTEAQMSISSGSSGHGGGMVTSALLCWAVALVFLSLTRVPVVQGDCWLIEGEKGFVWLAI
CSQNQPPYENIPMHINSTIVDLRLNENKIRSIHFSSLSRFGNLTYLNLTKNDISYIEDGA
FSAQFNLQVLQIGFNKMRNLTEGMLRGLGRLQYLYLQANLIETVTANAFWECPAIENIDL
SMNRIQQLDGATFRGLSKLTTCELYANPFSCSCELLGFLRWLVAFPNRTSERMACDTPAG
FSGYSLLSQNPRMPRYRNAFHALSSVCTDDNVTPYLHGSPDGGIPTRLPDLSPCGLDDCS
SGTFPEETISISPTFLDSDARPIMKLKEVTHTSAVITIQIPNPFRKMYILVLYNNSFFTD
IQNLKSQREEIELQNLKSHTNYTYCVASIRNSLRFNHTCLTISTGPRTLQERVVNDSTAT
HYIMTILGCLFGMLIVLGVVVHCLRKRKKQDEPKSKKLEKMKRNLIEMKYGTELEQGTIS
QLSQKQILSAGETVQRMPYLPAASDTDQYKMQEISGTPKTTKGNYMEVRSEQPERSIREC
SVPPSETSQGSVAEISTIAKEVDKVNQIINNCIDALKSDSTSFQTAKSGAVSTAEPQLVL
ISEHPPGKSGFLSPVYKGGYHHPLQRHHSMDAPQKRASTSSSGSLRSPRSFRSEGAYRSE
SKYIEKASPQDESGIITVTPAAAILRAEAERIRQYSEHRHSYPGPHHIEESIEVSGSRKS
SILEPLTRSRPRELSYSQLSPQYHNLSGYSSPEYSCRPSLSLWERFKLHRKRHRDEEEYI
AAGHALRRKVQFAKDEDLHDILDYWKGVSAQQKS-
Mutant Protein Sequence
MTEAQMSISSGSSGHGGGMVTSALLCCGSCVPLTDKGVCCPG-In Situ Probe Sequence
GGACATGGAGGAGGAATGGTGACCAGTGCCTTACTTTGCTGGGCTGTGGCTCTTGTGTTCCTCTCACTGACAAGGGTGCCTGTTGTCCAGGGTGATTGCTGGCTGATTGAAGGGGAAAAGGGATTTGTGTGGTTGGCAATTTGCAGCCAGAACCAGCCCCCCTATGAAAACATCCCAATGCATATTAACAGCACAATTGTTGACCTGCGCCTCAATGAGAACAAGATTCGAAGCATCCATTTTTCTTCCCTTAGCCGCTTTGGCAATCTCACATACCTGAATCTAACAAAGAATGACATCTCTTACATAGAAGATGGGGCTTTCTCTGCTCAATTTAATCTCCAAGTGCTGCAGATTGGCTTTAACAAAATGAGGAACTTGACTGAGGGAATGTTAAGAGGTTTGGGACGCTTACAGTACCTCTATCTTCAGGCAAACCTGATCGAGACCGTCACAGCCAATGCCTTTTGGGAATGTCCTGCGATAGAAAACATCGACCTCTCCATGAACAGGATTCAGCAGCTTGATGGCGCTACATTCCGTGGTCTATCAAAGCTTACGACGTGCGAGCTCTACGCTAACCCTTTCAGCTGCTCATGTGAGCTGTTGGGATTCTTGCGCTGGCTTGTTGCGTTCCCAAACCGGACAAGTGAGAGGATGGCGTGTGATACACCTGCCGGCTTTTCAGGCTACAGCCTCTTGAGTCAGAACCCTAGGATGCCCAGATATCGAAATGCTTTCCATGCCCTCTCTTCAGTCTGCACCGATGACAATGTGACTCCGTATCTTCATGGATCTCCAGATGGAGGCATTCCAACGAGGTTGCCTGATTTGAGTCCTTGCGGACTGGATGACTGTTCCTCTGGGACATTTCCAGAGGAGACAATTAGCATCAGCCCCACCTTTCTGGMutant Genotyping Size
499Ribosome Profiling Development
Behavior Data
Behavior Data Summary
NoneBehavior Data Description 1
Dark flash block 1 start Merged section Pvalue = 0.00708 14 homhet vs 20 homhom ribgraph_mean_ribbon_fullboutdata_dpix_a0darkflash103.pngBehavior Data Graph 1
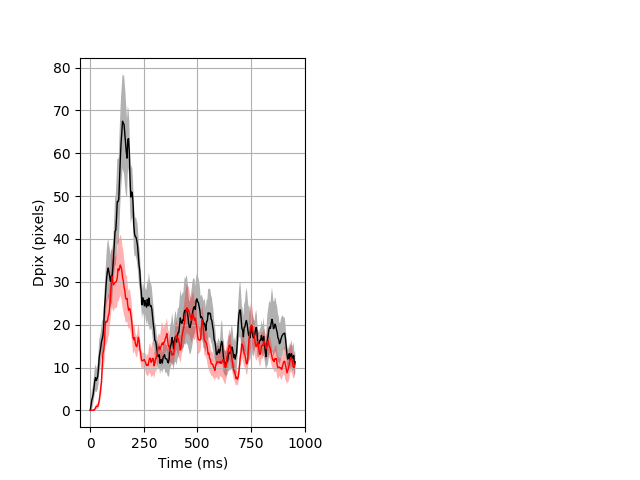
Behavior Data Description 2
Day all prepulse tap Merged section Pvalue = 0.0165 26 hethet vs 23 homhom ribgraph_mean_ribbon_fullboutdata_dpix_dayprepulseinhibition100d.pngBehavior Data Graph 2
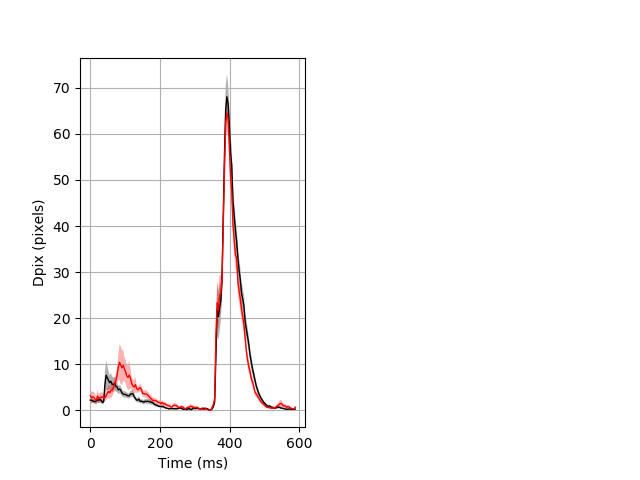
Behavior Data Description 3
Features of movement entire protocol Graph Pvalue = 0.00215 Merged section Pvalue = not significant might not repeat 26 hethet vs 24 hethom ribgraph_mean_ribbonbout_aveboutdisp_10min_combo.pngBehavior Data Graph 3
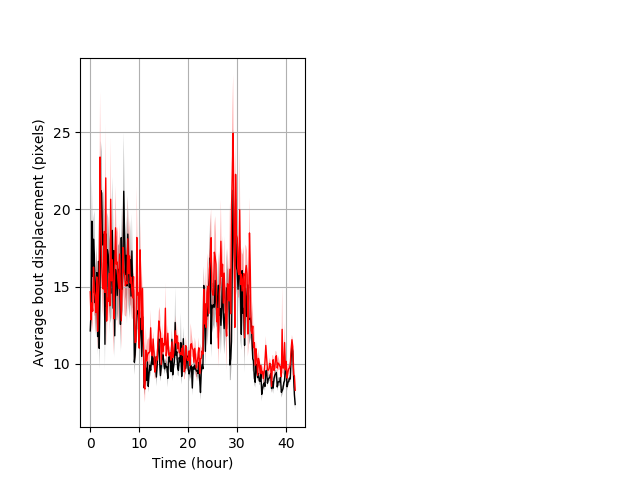
Behavior Data Description 4
Frequency of movement entire protocol Graph Pvalue = 0.0461 Merged section Pvalue = not significant 14 homhet vs 20 homhom ribgraph_mean_ribbonbout_dpixnumberofbouts_10min_combo.pngBehavior Data Graph 4
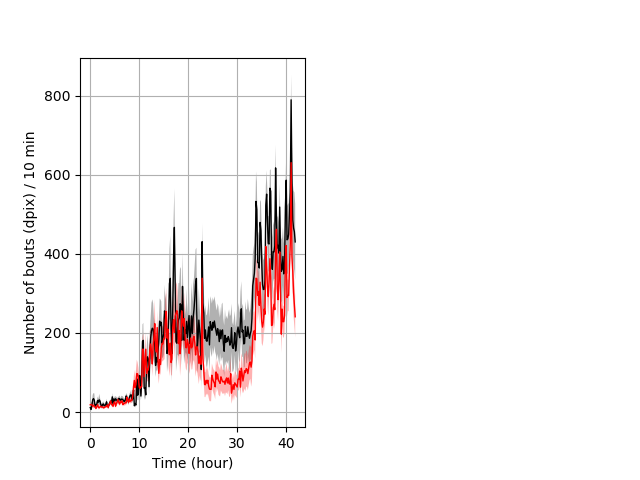
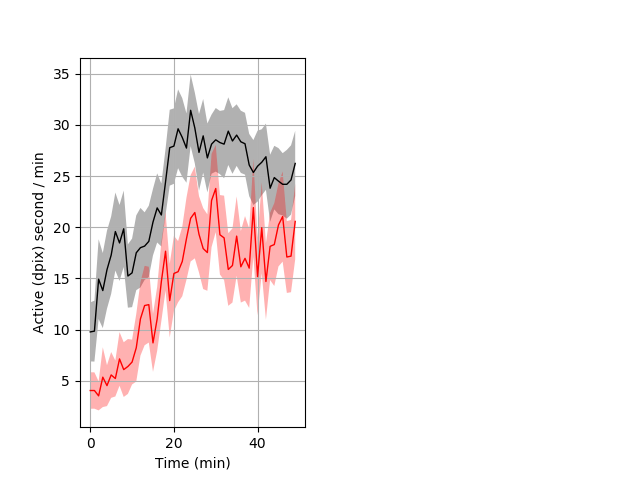
Behavior Data Description 5
Location in well entire protocol Graph Pvalue = 0.00379 Merged section Pvalue = 0.0064 17 hethom vs 20 homhom ribgraph_mean_ribbonbout_averhofrac_10min_combo.pngBehavior Data Graph 5
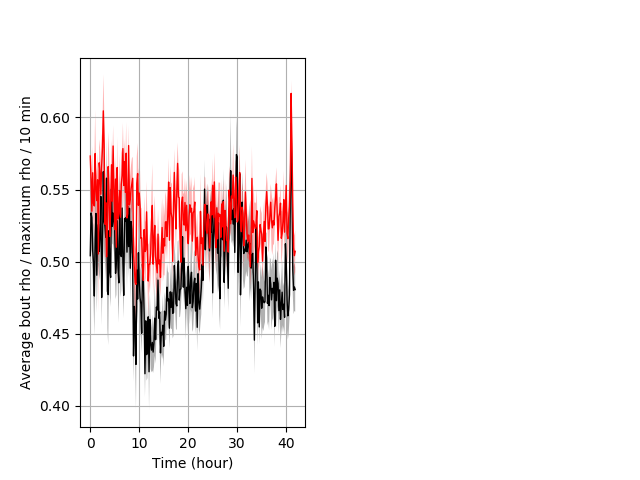
Behavior Data Description 6
Dark flash Graph Pvalue = 0.00136 20 hethet vs 20 homhom boxgraph_pd__ribgraph_mean_ribbon_displacement_dist_ddarkflash103.pngBehavior Data Graph 6
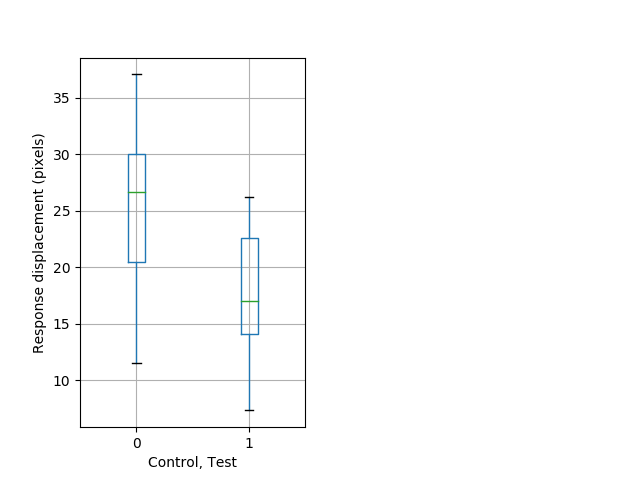
Behavior Data Description 7
Features of movement Graph Pvalue = 0.001 14 homhet vs 20 homhom ribgraph_mean_ribbonbout_aveboutvel_min_day2night2.pngBehavior Data Graph 7
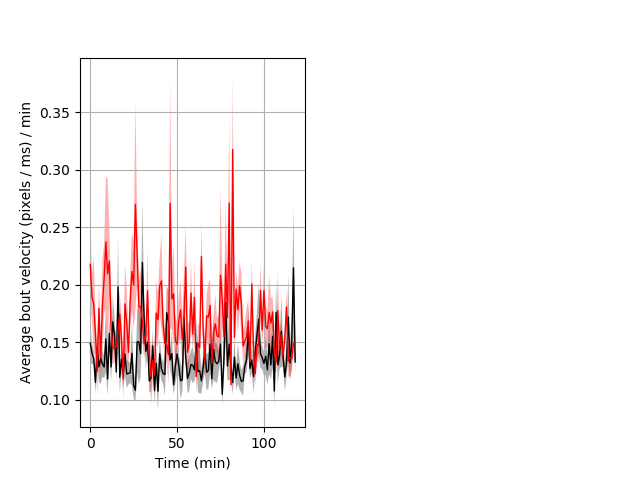
Behavior Data Description 8
Frequency of movement Graph Pvalue = 0.002 26 hethet vs 23 homhom ribgraph_mean_ribbon_dpixsecper_min_day2morning.pngBehavior Data Graph 8
Behavior Data Description 9
Light flash Graph Pvalue = 1.77e-05 31 hethet vs 19 homhet boxgraph_pd__ribgraph_mean_ribbon_peakdpix_lightflash104.pngBehavior Data Graph 9
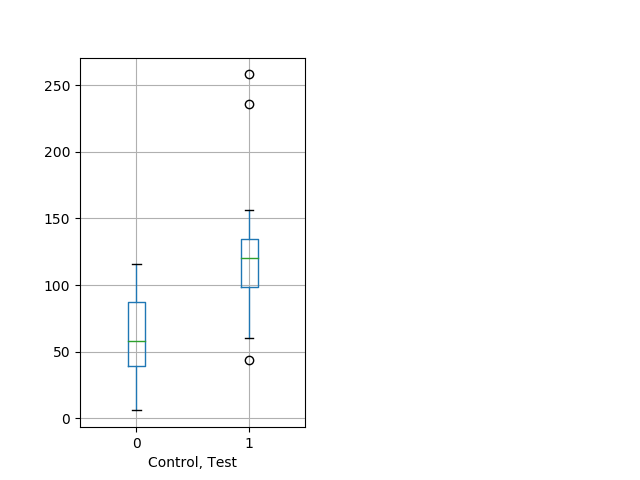
Behavior Data Description 10
Location in well Graph Pvalue = 7.44e-05 17 hethom vs 20 homhom ribgraph_mean_ribbonbout_centerfrac_10min_combo.pngBehavior Data Graph 10
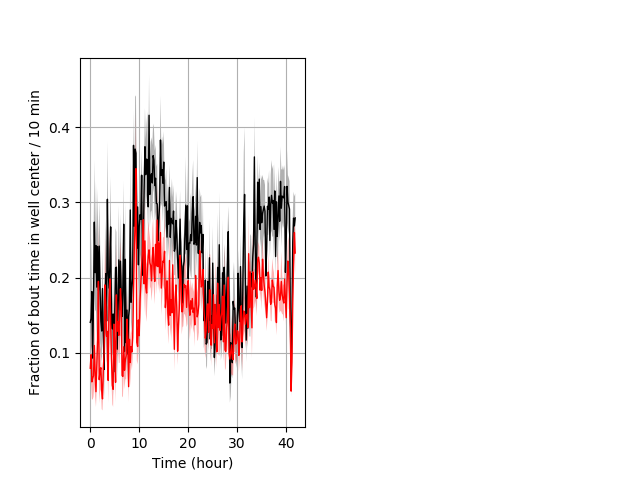
Behavior Data Description 11
PPI tap Graph Pvalue = 0.00193 31 hethet vs 25 hethom boxgraph_pd__ribgraph_mean_ribbon_freqresponse_dpix_shortnightprepulseinhibition100b.pngBehavior Data Graph 11
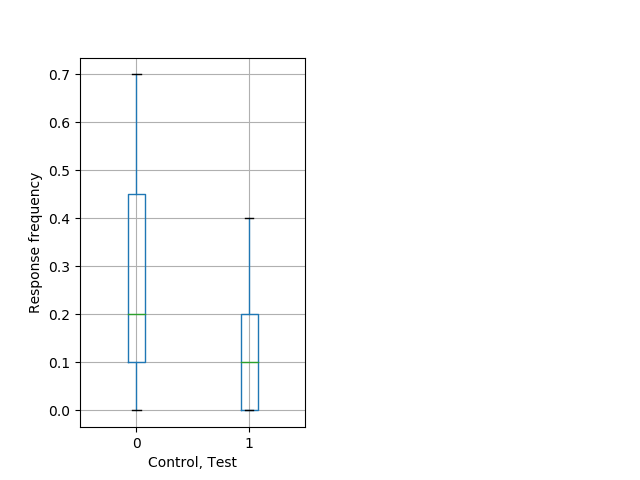
Behavior Data Description 12
Strong tap Graph Pvalue = 0.00505 20 hethet vs 17 hethom boxgraph_pd__ribgraph_mean_ribbon_totaldistanceresponse_dist_nightprepulseinhibition102.pngBehavior Data Graph 12
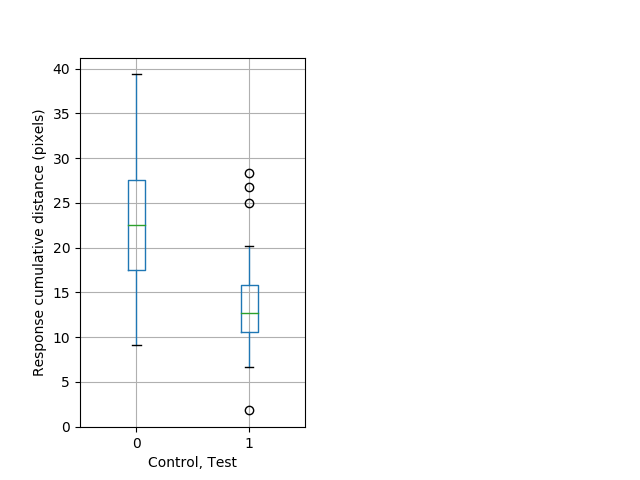
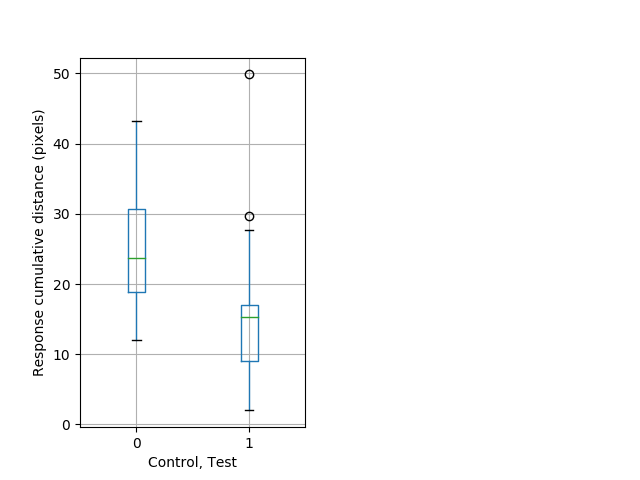
Behavior Data Description 13
Tap habituation Graph Pvalue = 0.00546 24 hethom vs 23 homhom boxgraph_pd__ribgraph_mean_ribbon_freqresponse_dpix_bdaytaphab1.pngBehavior Data Graph 13
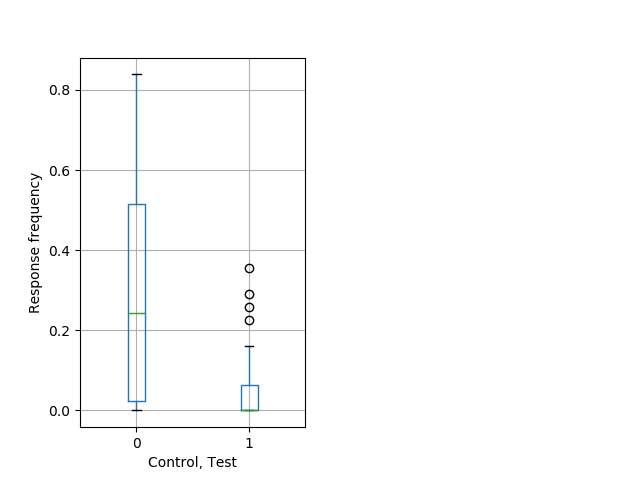
Behavior Data Description 14
Weak tap Graph Pvalue = 0.000815 31 hethet vs 19 homhet boxgraph_pd__ribgraph_mean_ribbon_totaldistanceresponse_dist_dayprepulseinhibition101.pngBehavior Data Graph 14