Locus Rank
7Gene
Id
128Gene Name
MAD1L1Duplicated
FalseMaternal
TrueGene Not Within Locus (Nearby
FalseGene Description
Mouse Phenotype
Death, etcAdditional Image
Allen Brain
Allen Regions
Zfin In Situ
Zfin Brain Areas
Published Zebrafish Pehnotype
Not In Allen Brain
TrueZFIN Link
http://zfin.org/ZDB-GENE-040426-1081Allen Link
http://mouse.brain-map.org/gene/show/16890More Additional Images
Papers
Locus Rank
Locus Rank
7Genes in Region
MAD1L1 (only gene in locus), and CHST12, SNX8, FTSJ2, EIF3B, NUDT1, and ELFN1 nearby locus.Associated Snps
GWAS Region (hg19)
chr7:1896096-2190096Genes Skipping
SNX8 - no brain expression, seems unlikely based on pubmed; NUDT1 - also not allen brain, and nothing about neurons in any paper; FTSJ2 - cell cycle, dna repair; EIF3B - essential roles in protein synthesisRicopili Plot Surrounding Area
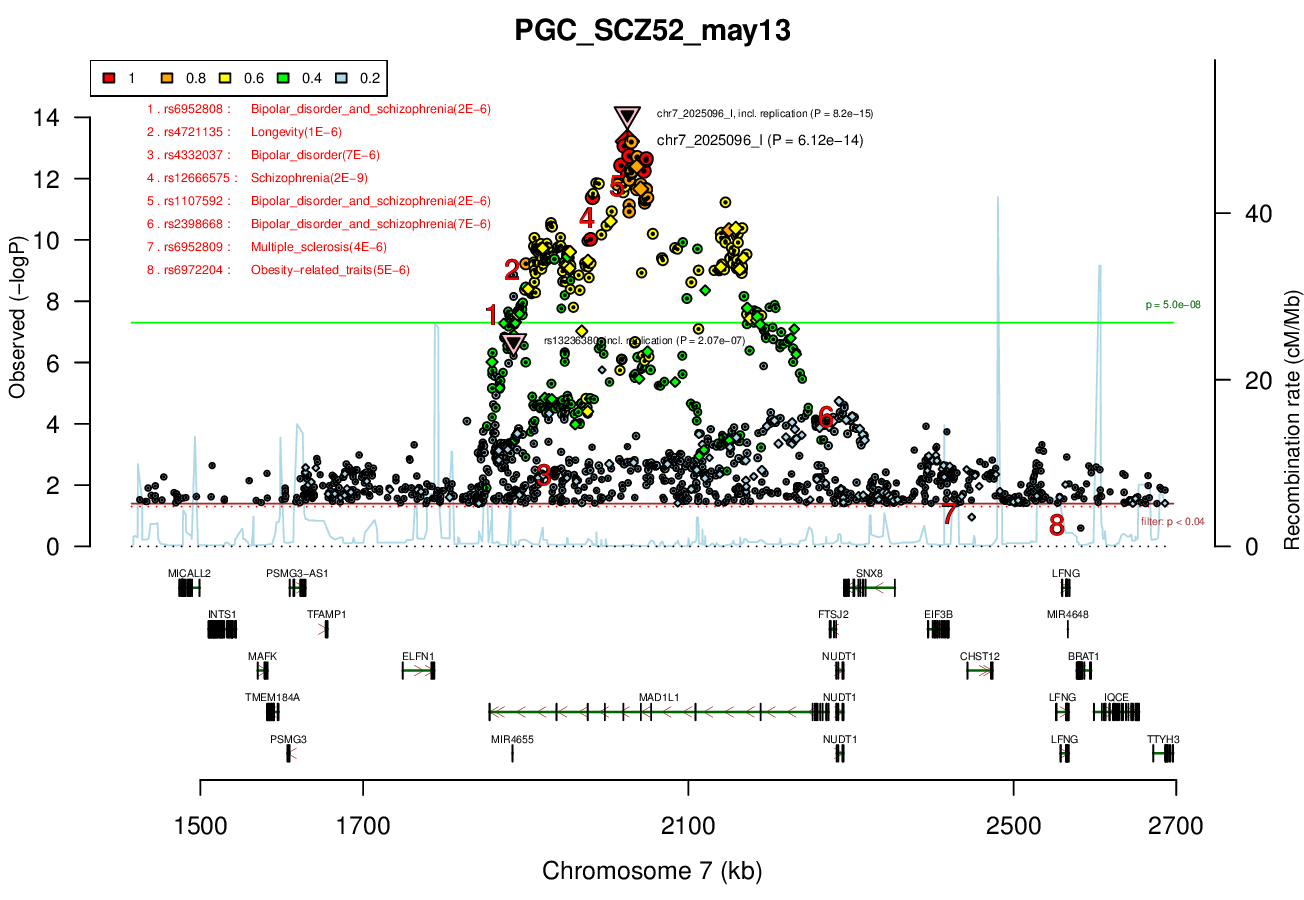
Distance On Each Side Of Locus In Above Plot
0.5 MBRicopili Plot
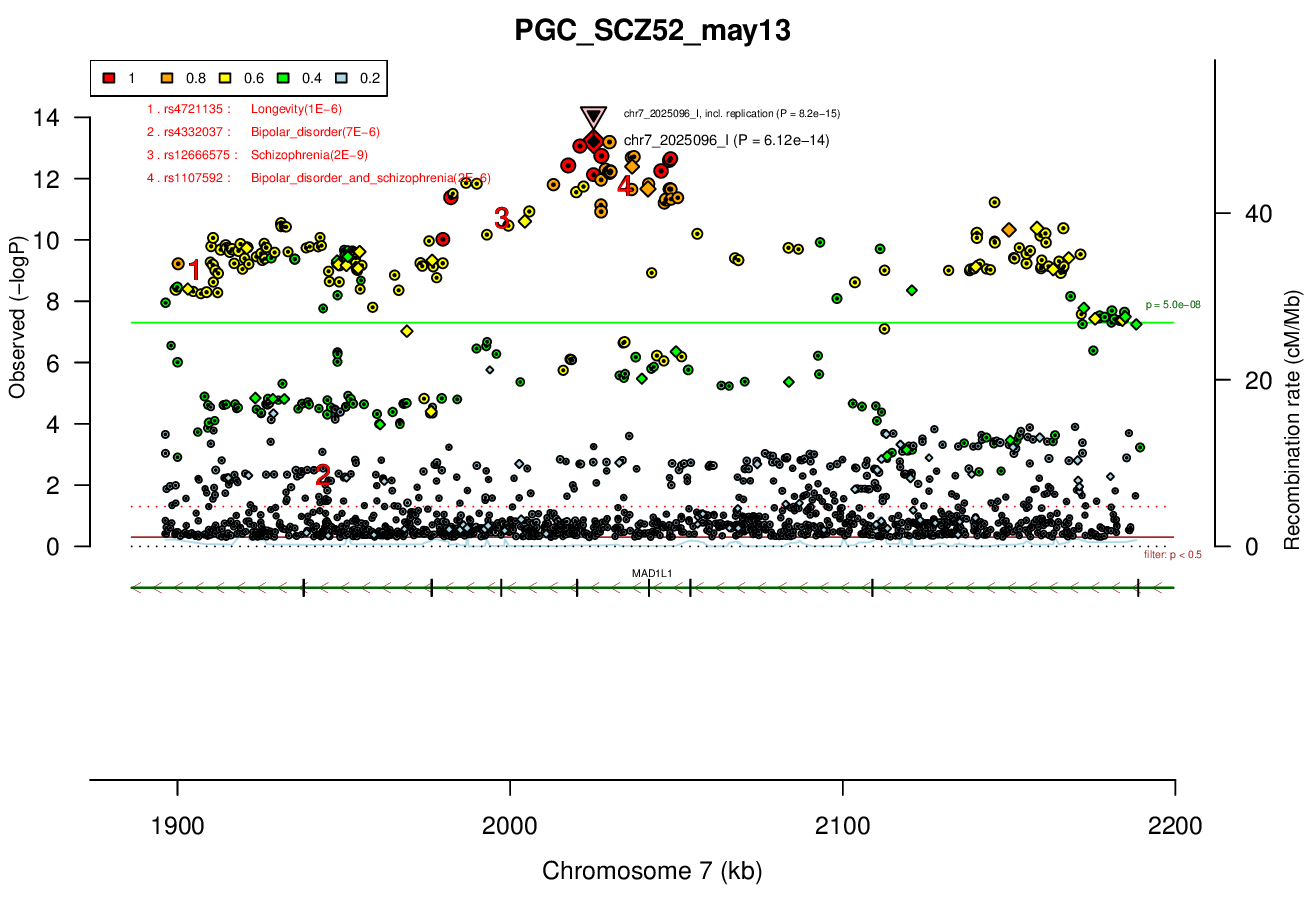
Gwas And Surrounding Region Pic
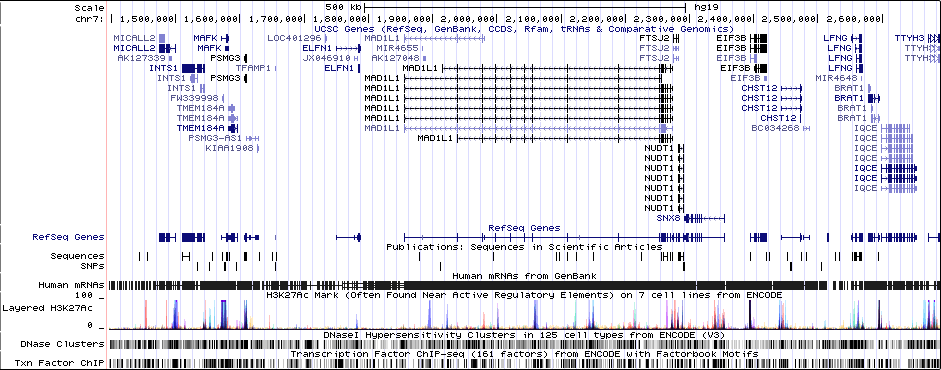
Gwas Region Pic
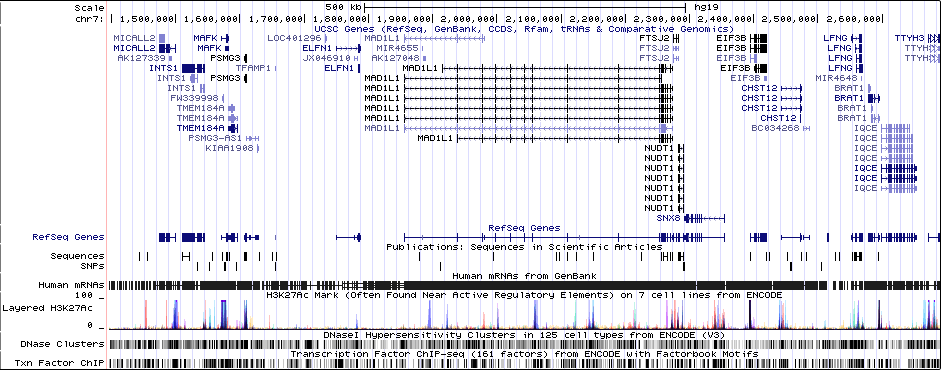
Sequence 1
Ensembl Gene Name
mad1l1Harvard Allele
a296Mutation Area - WT DNA Sequence
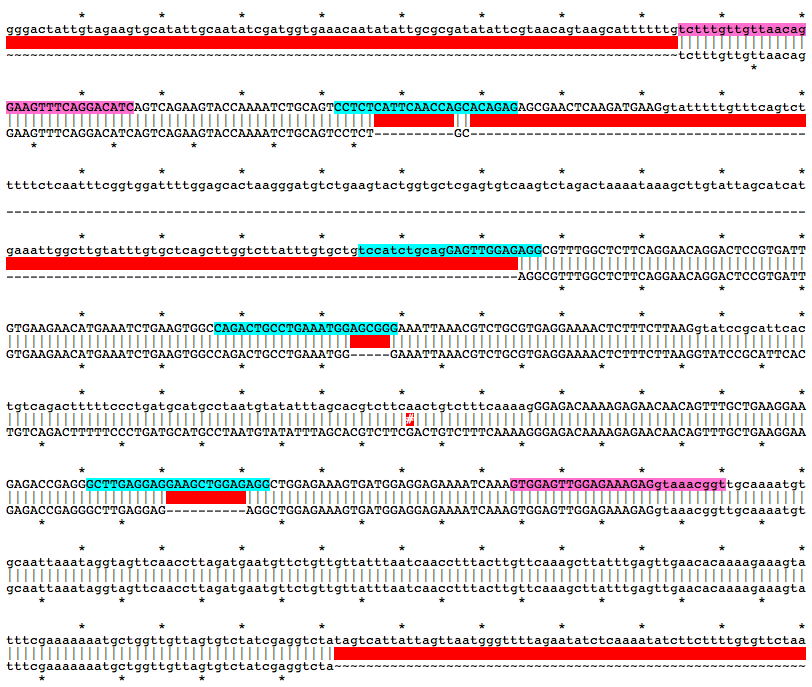
tctttgttgttaacagGAAGTTTCAGGACATCAGTCAGAAGTACCAAAATCTGCAGTCCTCTCATTCAACCAGCACAGAGAGCGAACTCAAGATGAAGgtatttttgtttcagtctttttctcaatttcggtggattttggagcactaagggatgtctgaagtactggtgctcgagtgtcaagtctagactaaaataaagcttgtattagcatcatgaaattggcttgtatttgtgctcagcttggtcttatttgtgctgtccatctgcagGAGTTGGAGAGGCGTTTGGCTCTTCAGGAACAGGACTCCGTGATTGTGAAGAACATGAAATCTGAAGTGGCCAGACTGCCTGAAATGGAGCGGGAAATTAAACGTCTGCGTGAGGAAAACTCTTTCTTAAGgtatccgcattcactgtcagactttttccctgatgcatgcctaatgtatatttagcacgtcttcaactgtctttcaaaagGGAGACAAAAGAGAACAACAGTTTGCTGAAGGAAGAGACCGAGGGCTTGAGGAGGAAGCTGGAGAGGCTGGAGAAAGTGATGGAGGAGAAAATCAAAGTGGAGTTGGAGAAAGAGgtaaacggtMutation Area - Mutant DNA Sequence
tctttgttgttaacagGAAGTTTCAGGACATCAGTCAGAAGTACCAAAATCTGCAGTCCTCTGCAGGCGTTTGGCTCTTCAGGAACAGGACTCCGTGATTGTGAAGAACATGAAATCTGAAGTGGCCAGACTGCCTGAAATGGGAAATTAAACGTCTGCGTGAGGAAAACTCTTTCTTAAGGTATCCGCATTCACTGTCAGACTTTTTCCCTGATGCATGCCTAATGTATATTTAGCACGTCTTCGACTGTCTTTCAAAAGGGAGACAAAAGAGAACAACAGTTTGCTGAAGGAAGAGACCGAGGGCTTGAGGAGAGGCTGGAGAAAGTGATGGAGGAGAAAATCAAAGTGGAGTTGGAGAAAGAGgtaaacggttgcaaaatgtgcaattaaataggtagttcaaccttagatgaatgttctgttgttatttaatcaacctttacttgttcaaagcttatttgagttgaacacaaaagaaagtatttcgaaaaaaatgctggttgttagtgtctatcgaggtctaGuide RNA Target Sites
CTCTGTGCTGGTTGAATGAGAGG
tccatctgcagGAGTTGGAGAGG
CAGACTGCCTGAAATGGAGCGGG
GCTTGAGGAGGAAGCTGGAGAGG
Genotyping Primers
f, tctttgttgttaacagGAAGTTTCAGGACATC
r, accgtttacCTCTTTCTCCAACTCCAC
Alignment of WT and Mutant Sequences w/ gRNA Targets (cyan) and Genotyping Primers (pink) Shown on WT Sequence
Allele
216bpD,5bpD,10bpDWT Genotyping Size
606WT Protein Sequence
MDDIEDDTTIFSTLKSFKSLISCPERSLLETEPAYGRSDLQKLYTKRVELEEAAERVRSH
TSLIQLTQEKQQMELSHKRARIELEKEAHSSSRDLQREIDRNRDLQMKIRRLEEREEKAN
QALNEQMENNKALKRNLEELHKKANEKDSKLTEAEQIISELKDETRRLGQQIQIQESKFS
TQNLEIQALQEQLDVQRKKFQDISQKYQNLQSSHSTSTESELKMKELERRLALQEQDSVI
VKNMKSEVARLPEMEREIKRLREENSFLRRETKENNSLLKEETEGLRRKLERLEKVMEEK
IKVELEKEKMERELQAWENIGQATGLNIRKPEDLSREVIQIQQRELNFKQQNYSLNSSVR
SVEKARSELLTEIAQLRAKAQEEQKKRENQDSLVRRLQKRVLLLTKERDGMRAILESYDS
ELATSEYSPQLTLRVKEAEDMLQKVQAHNAEMETQLSKAQEEAGSFKLQAQMVAAELEAL
KEQQVSNAERSSSVTAEEISSLRQKIEELEAERQRLEEQNNILEMRLERHNLQGDYDPVK
TKVVHLQMNPTSMAKQQRADEVEQLRVECQRLRDRLRKIEVAGGMTTDDTTLIIPPSQEI
LDLRKQMESAELKNQRLKEVFQKKIQEFRTACYVLTGYQIDITVENQYRLTSVYAEHMED
SLLFKSTGPVGSGSMQLLETDFSRTLTGLVDLHLFHQKSIPVFLSAVTIELFSRQTVA-
Mutant Protein Sequence
MDDIEDDTTIFSTLKSFKSLISCPERSLLETEPAYGRSDLQKLYTKRVELEEAAERVRSH
TSLIQLTQEKQQMELSHKRARIELEKEAHSSSRDLQREIDRNRDLQMKIRRLEEREEKAN
QALNEQMENNKALKRNLEELHKKANEKDSKLTEAEQIISELKDETRRLGQQIQIQESKFS
TQNLEIQALQEQLDVQRKKFQDISQKYQNLQSSAGVWLFRNRTP-
Mutant Genotyping Size
536Ribosome Profiling Development
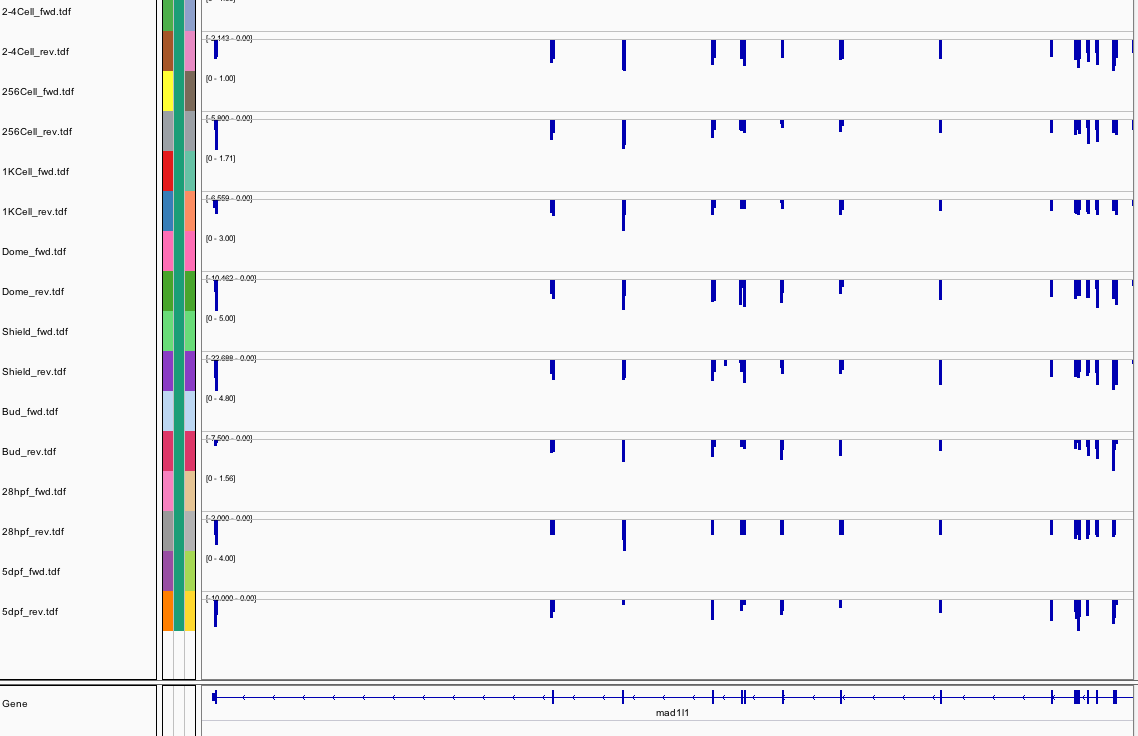
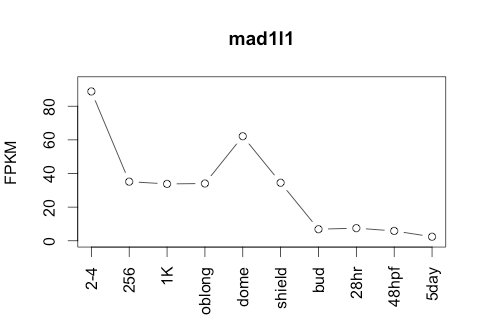
Transcript Plot
Behavior Data
Behavior Data Summary
NoneBehavior Data Description 1
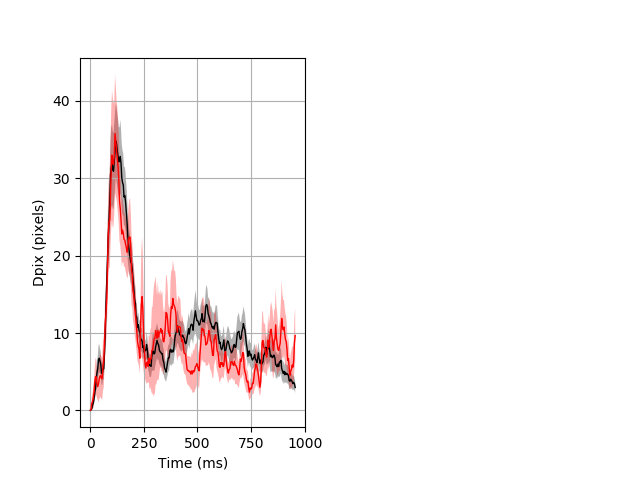
Dark flash block 1 start Merged section Pvalue = 0.0316 47 hetandwt vs 16 hom ribgraph_mean_ribbon_fullboutdata_dpix_a0darkflash103.pngBehavior Data Graph 1
Behavior Data Description 2
Day all prepulse tap Merged section Pvalue = not significant 23 wt vs 20 hom ribgraph_mean_ribbon_fullboutdata_dpix_dayprepulseinhibition100d.pngBehavior Data Graph 2
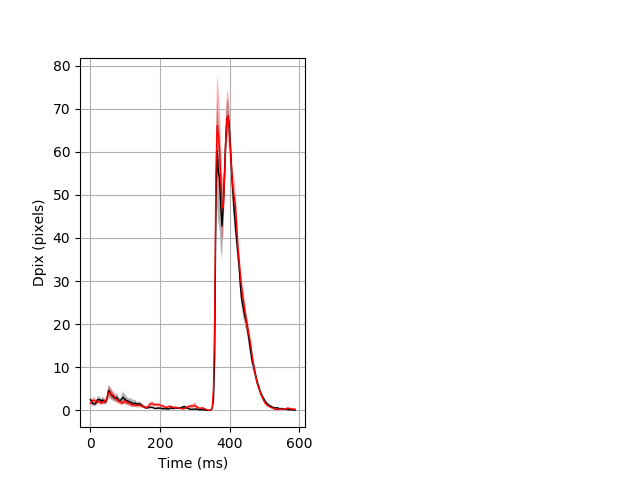
Behavior Data Description 3
Features of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 14 wt vs 16 hom ribgraph_mean_ribbonbout_aveboutdisp_10min_combo.pngBehavior Data Graph 3
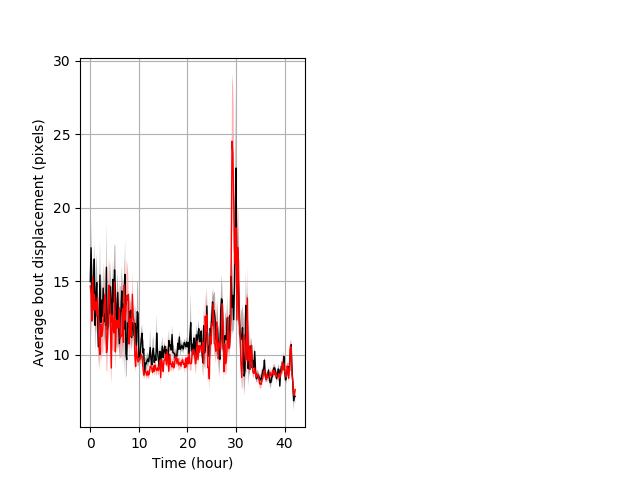
Behavior Data Description 4
Frequency of movement entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 23 wt vs 20 hom ribgraph_mean_ribbonbout_dpixnumberofbouts_10min_combo.pngBehavior Data Graph 4
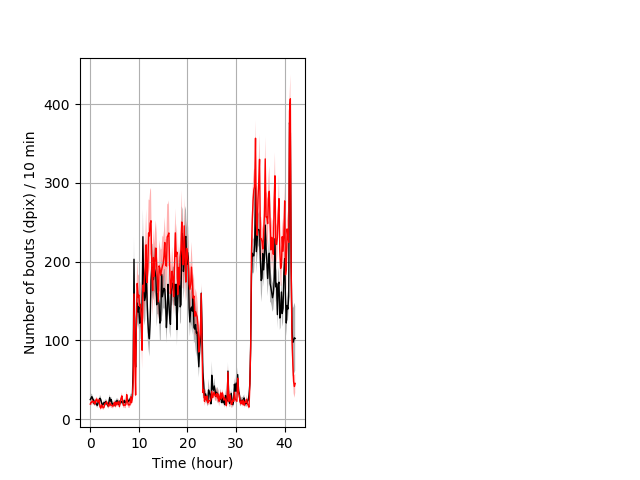
Behavior Data Description 5
Location in well entire protocol Graph Pvalue = not significant Merged section Pvalue = not significant 14 wt vs 16 hom ribgraph_mean_ribbonbout_averhofrac_10min_combo.pngBehavior Data Graph 5
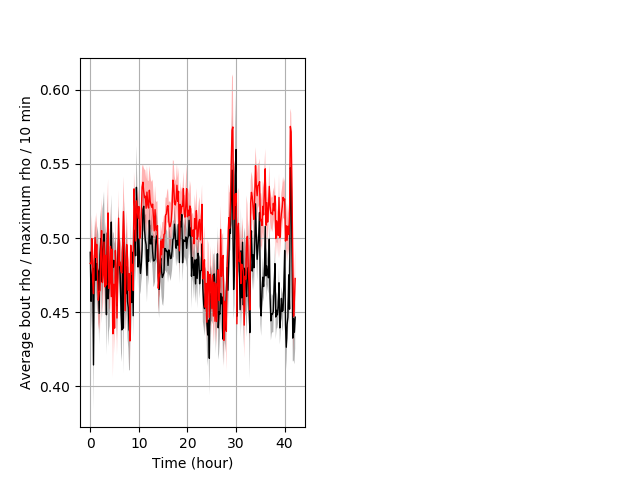
Behavior Data Description 6
Strong tap Graph Pvalue = 0.00748 14 wt vs 16 hom boxgraph_pd__bigmovesribgraph_mean_ribbon_polygonarea_dist_nightprepulseinhibition102.pngBehavior Data Graph 6
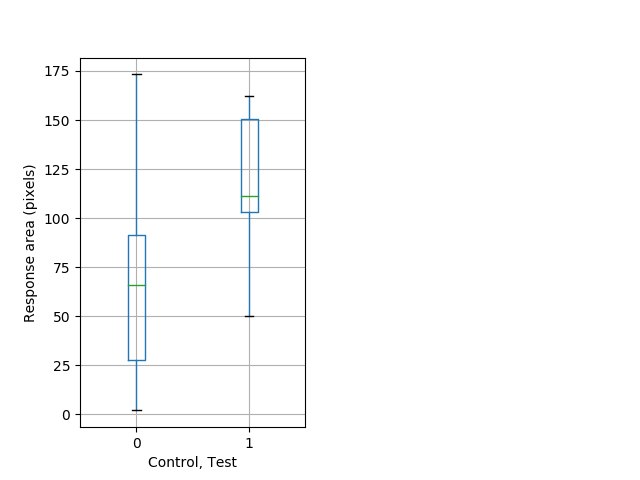
Behavior Data Description 7
Tap habituation Graph Pvalue = 0.00649 14 wt vs 33 het boxgraph_pd__ribgraph_mean_ribbon_peakdpix_adaytaphab102.pngBehavior Data Graph 7
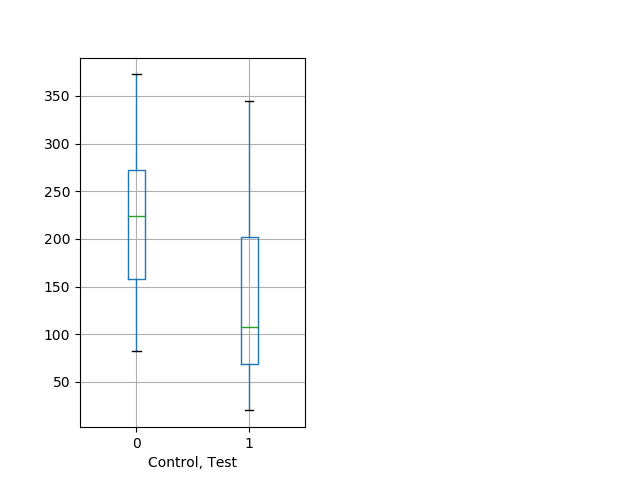
Behavior Data Description 8
Weak tap Graph Pvalue = 0.0116 69 hetandwt vs 20 hom boxgraph_pd__smallmovesribgraph_mean_ribbon_timeresponse_dpix_shortnightprepulseinhibition101.pngBehavior Data Graph 8
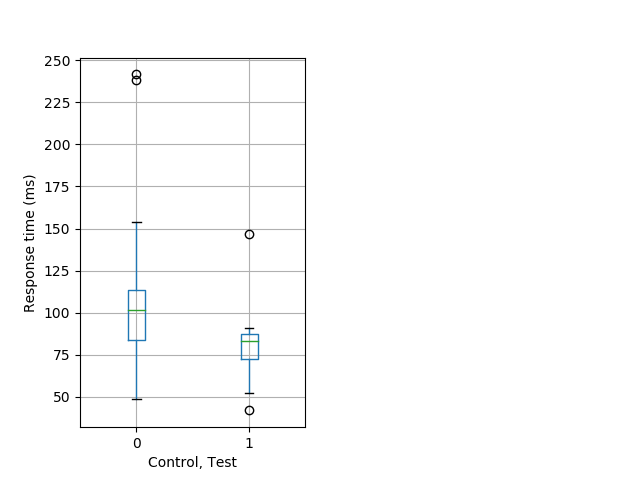
Behavior Data Description 9
NoneBehavior Data Graph 9
NoneBehavior Data Description 10
NoneBehavior Data Graph 10
NoneBehavior Data Description 11
NoneBehavior Data Graph 11
NoneBehavior Data Description 12
NoneBehavior Data Graph 12
NoneBehavior Data Description 13
NoneBehavior Data Graph 13
NoneBehavior Data Description 14
NoneBehavior Data Graph 14
None